Rethinking Tautomer Representation
If ever there was a perennial problem in cheminformatics, it would be tautomerism. Dealing with it consistently and in a chemically relevant way is much harder than appearances might suggest. The problem is multi-faceted, requiring not just a rigorous definition of a slippery concept, but methods to pinpoint molecular features leading to tautomerism. These problems get most of the attention, but tautomer representation, a problem of equal importance, gets almost none. This article takes a closer look at tautomer representation and proposes an unconventional approach.
Proposal
Tautomerism complicates the comparison of molecular representations for equivalence. Here'a a procedure that solves this problem:
- Find all tautomeric groups. A tautomeric group is a set of connected atoms and bonds capable of giving tautomers.
- Within each tautomeric group demote all double bonds. Also demote single bonds to hydrogen.
- Compare the transformed representations for equivalence.
Demotion is a procedure in which the order of a bond is reduced by one without changing the constitution of the parent molecular graph. A triple bond becomes a double bond, a double bond becomes a single bond, and a single bond is deleted altogether. The details are described in the next section.
For now, consider the example of 4-methylimidazole. Imagine that an organization's business rules consider imidazole tautomers to be equivalent. However, direct comparison of the tautomers of 4-methylimidazole erroneously finds them to be non-equivalent. The expected result can be obtained via the procedure outlined above.
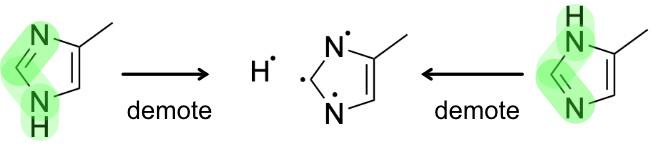
The remainder of this article describes the procedure in detail.
Bond Demotion
Demotion reduces a bond's order by one while adjusting atomic properties to maintain the original constitution. Any bond can be demoted, but this article focuses on use with tautomer representation.
Demotion consists of the following steps:
- If either terminal atom uses implicit hydrogen counting, disable it and set a virtual hydrogen count.
- Reduce bond order by one. If the resulting bond order is zero, delete the bond.
- Remove conformational descriptors, if any, from the demoted bond.
The following examples (expressed as Balsa strings) illustrate demotion:
*N[H]→*[NH].[H]*[C@](F)(Cl)[H]→*[C](F)(Cl).[H]*/C=C/*→*[CH][CH]**C#C*→*[C]=[C]*
The VB Model
Most widely-used small molecule representation build on the Valence Bond (VB) model. This model views a covalent bond as composed of two atoms and 2n shared electrons, where n is an integer greater than zero. In other words, a covalent bond is little more than an edge of a molecular graph labeled with an electron count or bond order.
The VB model fits enough accumulated experimental evidence to still be relevant after a century of use. But it's far from perfect.
Electron Delocalization
As George Box famously noted, "All models are wrong, but some are useful.". Despite its enormous success, the VB model is wrong in some pretty spectacular ways.
The most glaring example is electron delocalization ("delocalization"). A previous article defined delocalization as "…the phenomenon in which the electrons of a VB representation can be distributed over more than one bond, among two or more atoms, or across some combination of atoms and bonds."
Consider 1,2-difluorobenzene. Two VB representations are possible, but they are almost universally considered equivalent. The diagrams depict six electrons localized across three double bond. Nevertheless, the experimental evidence overwhelmingly supports a view in which those six electrons are shared equally among the six ring atoms.
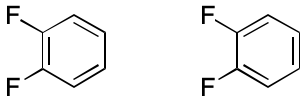
Delocalization of electrons around an alternating cyclic path, as in 1,2-difluorobenzene, is an important failure mode for the VB model. But there are others.
What is Tautomerism, Anyway?
Despite its importance to chemistry and related fields, there's a lot of disagreement about what exactly tautomerism is. Fortunately, there's also a nugget of agreement as well.
Perhaps the most influential tautomer definition is the one embedded within the InChI software. The 2015 description defines tautomerism in InChI as one of two possible structural transformations:
M=Q-ZH↔MH-Q=ZM=Q-Z⊖↔M⊖-Q=Z
where M and Z are elements in one of several possible valence states, Q is a neutral atom, and H is any isotope of hydrogen.
InChI's approach was clearly influenced by the definition of tautomerism developed at CAS in the late 1970s, although the connection could have been made more clear. The CAS system is based on "… algorithmically recognizing tautomeric and alternating bond structures, replacing the explicit single and double bonds with special normalized bonds, and associating the migrating tautomeric hydrogen with groups of atoms rather than just single atoms." Both Standard InChI and CAS disregard keto-enol tautomerism, allowing each form to be treated as a distinct entity.
The CAS authors define tautomerism as: "… a state of equilibrium in which a mobile group, typically a hydrogen, migrates between atoms with concurrent changes in bonding. The basic generic tautomeric structure is:"
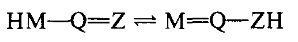
The CAS definition also supported the generalized tautomeric structural motif in which double bonds intervene between M and Q, and in which branching can occur.
For completeness, two reviews published in 2010 are often cited in the context of tautomerism:
- So you think you understand tautomerism?. Many examples of tautomerism are presented, and approaches to tautomerism are classified as either "local" or "global."
- Tautomerism in chemical information management systems. Summarizes the tautomer policies of more than two dozen software vendors.
Tautomerism is sometimes erroneously identified with hydrogen "motion" or "movement." Even the term "migration" conjures and image of atoms moving from one relative position in space to another. But as the definitions from CAS and InChI make clear, the coordinates of atoms are not only irrelevant but undefined in many cases.
A better term for what happens in tautomerism is rebonding. Rebonding changes the bond order between two atoms in a representation. The common thread among all tautomer definitions is the rebonding of one or more groups of atoms. This can take place with or without atomic motion. Cheminformatics often deals with representations that lack coordinates, or whose coordinates are merely aesthetic in nature. To require motion as part of a tautomer definition implies the existence of information that doesn't necessarily exist.
Bond demotion works as a technique for dealing with tautomerism because tautomerism is fundamentally a process of rebonding.
Tautomerism and Molecular Equivalence
Tautomerism is especially troublesome in the context of molecular equivalence ("equivalence"). Two representations are said to be equivalent if they encode the same molecule. An equivalence check is a fundamental operation when building molecule collections, which appear in a wide range of guises.
The example that started this article showed how tautomeric forms viewed as equivalent in one context can nevertheless be viewed as non-equivalent in other contexts. Tautomerism undermines the goal of building a collection of unique molecule by masking equivalence relationships. But this is is really just another form of the one with 1,2-difluorobenzene.The root cause in both cases was electron delocalization.
False non-equivalence signals like those presented by 1,2,-difluorobenzene can be address by ignoring bond order in favor of atomic valence (aka "bond order sum"). Doing so removes bond order from consideration, thereby suppressing false negatives.
This shift in perspective can be approached from several directions. One of the simplest is to demote all double bonds. The result is a representation devoid of double bonds, yet nevertheless consistent with constitution of the original molecule. A useful side-effect of this approach is that it translates well to string canonicalization.
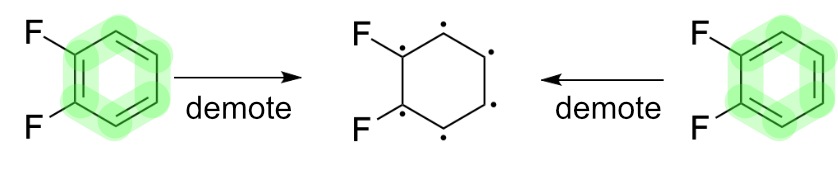
Bond demotion solves the problem in cases such as 1,2-difluorobenzene, but it also works for tautomerism.
What's the Actual Problem?
It's tempting to think of the delocalization problem as resulting from some missing capability. To account for bonds that are not single, double, or triple, a new bond type might be introduced. Indeed, a recurring issue with the V2000 molfile format is the misappropriation of bond type 4 for just this purpose. The biggest problem with adding bond types is the massive community investment (software, databases, validation, etc.) in representation systems that would not understand it.
Bond demotion offers a compatible path forward. Rather than adding a feature, one or more features can be selectively disabled — in this case bond order. The result is a representation that can be processed by existing software. A previous article explains the idea in more detail.
From this perspective, the problem with th VB model isn't lack of features. Rather, it's that indiscriminate use of certain features leads to overspecification. A double bond drawn between the fluorene-bearing carbons of 1,2,-difluorobenzene means something quite different than one drawn between the fluorine-bearing carbons of 1,2-difluoroethene.
Tautomerism as a Special Case of Electron Delocalization
Tautomerism can be viewed as a special case of electron delocalization, at least in the context of molecular equivalence comparisons. The distinguishing trait for tautomerism is the rebonding of single bonds. Under this model, the delocalization in both 1,2-difluorobenzene and 4-methylimidazole are merely two sides of the same coin.
The main practical advantage is to allow equal treatment of two structural phenomena that previously had to be treated differently.
Objections
An understandable objection to this approach invokes the concept of "energy barrier." The two valence bond forms of a molecule like 1,2-difluorobenzene exist in superposition with each other — without an energy barrier. In contrast, the two valence bond forms of 4-methylimidazole are potentially distinct species which can be observed and/or isolated. In other words, tautomeric forms are separated by an energy barrier.
Wikipedia captures this nuance with the following caveat:
Care should be taken not to confuse tautomers with depictions of "contributing structures" in chemical resonance. Tautomers are distinct chemical species that can be distinguished by their differing atomic connectivities, molecular geometries, and physicochemical and spectroscopic properties, whereas resonance forms are merely alternative Lewis structure (valence bond theory) depictions of a single chemical species, whose true structure is best described as the "average" of the idealized, hypothetical geometries implied by these resonance forms.
Granted, but I'd propose that the utility of this distinction depends on context. The valence bond model fails in either case, and in exactly the same way. No single VB representation fits observation. The valence bond model over-constrains electronic structure, leading to purely artifactual non-equivalence.
This line of thinking can be taken one step further by considering another similarity: electron delocalization always involves an estimation of energy barrier, whether tautomerism is involved or not. Consider "anti-aromatic" systems such as cyclobutadienes and cyclooctatetraenes. In principle the single and double bonds have different lengths and do not "rapidly" interconvert. Without knowing more about context, it's impossible to distinguish observable isomers from artifactual electron localization.
Finally, this proposal says nothing about how to identify tautomeric features. It merely offers some tools for dealing with the consequences of finding one.
Conclusion
Tautomerism and electron delocalization are often treated as separate phenomena. In many contexts, there's good reason to do so. However, molecular equivalence determination is not one of them. An alternative model is proposed that views tautomerism as a special case of electron delocalization. The same simple yet powerful tool, bond demotion, can then be applied uniformly regardless of the form electron delocalization takes.