Hydrogen Suppression in SMILES
SMILES is the most widely-used line notation in cheminformatics, and one of two standard information exchange formats. Like Molfile, SMILES supports hydrogen suppression, a method for representing monovalent hydrogens and associated bonds without explicitly encoding them within the molecular graph. But getting that system to work well requires clear, readily-available documentation. This article attempts to solve that problem.
Hydrogen Suppression
Hydrogen Suppression is the practice of omitting some or all hydrogen atoms from a molecular graph. Two forms are commonly encountered: implicit hydrogens and virtual hydrogens.
Implicit hydrogens disappear without a trace from the graph in which they are deleted. These deleted hydrogens and their bonds can be reconstituted only through the application of a set of rules referred to as a valence model. A valence model is a set of rules for assigning an implicit hydrogen count to an atom.
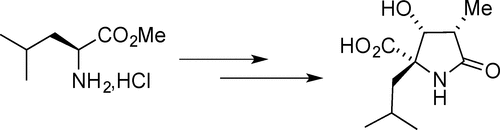
2D chemical structures offer a visual demonstration of implicit hydrogens. These drawings elide the hydrogens attached to most carbons. These hydrogens can be reconstituted with the simple rule that carbon has a target valence of four. To obtain a carbon atom's implicit hydrogen count, subtract its sum of bond orders from four.
In contrast, virtual hydrogens leave an imprint on the molecular graph from which they are deleted. This imprint takes the form of an atomic hydrogen count attribute. Unlike implicit hydrogens, virtual hydrogens can be reconstituted without a valence model. What you see is what you get. As a visual example, 2D chemical structures use virtual hydrogens to represent hydrogens suppressed on heteroatoms.
SMILES supports both virtual and implicit hydrogens. The system bears a close resemblance with the one used in 2D structure diagrams. Conveniently, SMILES clearly demarcates virtual and implicit hydrogen regimes with square brackets.
Bracket Atoms
A bracket atom is enclosed within the left and right square bracket characters ([ and ], respectively). Within brackets, SMILES uses virtual hydrogens exclusively. An optional hydrogen count is set by the character H followed by an optional count digit. If the digit is not present, then the count is one. If the leading H is not present, the count is zero.
For example, lithium metal would be represented as [Li]. No H appears within the brackets, therefore the atom has a virtual hydrogen count of zero. Similarly, lithium hydride would be represented as [LiH]. The utility of this convention extends beyond metals. Consider the methyl radical, which can be represented as [CH3], or a methyl carbene, which can be represented as [CH2]. No special rules and no lengthy documentation are required for these unusual valence states. What you see is what you get.
Bracket atoms aren't just for radicals, though. We could for example represent methane as [CH4]. However, this construct is so common and repetitive that SMILES offers a more concise alternative.
Bare Atoms
Atoms not enclosed within brackets use implicit hydrogens. Hydrogen counts on these atoms are under the control of a valence model. The only way to override this behavior is to enclose the atom within brackets.
The main advantage of this system is brevity. Automatically encoding hydrogen atom counts of the most common elements through a valence model saves a lot of space. For example, methane is represented as C, ammonia as N, water as O, and so on. Aggregates involving these atoms become quite easy to encode and read. For example, isopropanol can be represented as OC(C)C.
Of course, there's a catch. To correctly reconstitute the hydrogens in a SMILES using bare atoms, we need to obtain, correctly interpret, and correctly implement a valence model.
Valence Model
The goal of the SMILES valence model is to assign an implicit hydrogen count to any bare atom. The procedure is as follows:
- Determine the target valence for the atom. A target valence is the maximum number of bonding electrons an atom can contribute.
- Find the atom's bond order sum, also known as valence.
- Implicit hydrogen count equals target valence minus valence.
Target valences are identified by reference to the following table:
| Element | TargetValences |
|---|---|
| B | 3 |
| C | 4 |
| N | 3, 5 |
| O | 2 |
| P | 3, 5 |
| S | 2, 4, 6 |
| Halogens | 1 |
Consider, for example, propene (CC=C). We'd like to compute the implicit hydrogen count for the center atom. Referring to the table, carbon supports one target valence of four. The atom's valence (sum of bond orders) is three (2 + 1). Therefore, the implicit hydrogen count is one (4 - 3).
Elements supporting multiple target valences use the one closest to, but not less than, the atom's valence. For example, consider dimethylphosphite (P(=O)(OC)OC). The valence of the phosphorous atom is 4 (2 + 1 + 1). Phosphorous supports two target valences (3 and 5). The target valence closest to, but not less than 4 is five. Therefore, the implicit hydrogen count for phosphorous in dimethylphosphite is 1 (5 - 4).
If the molecule were trimethylphosphite (P(=O)(OC)(OC)OC), then the target valence would be five and the implicit hydrogen count would be zero.
Another Perspective
A shift in perspective offers insights into the similarities between the two hydrogen suppression models in SMILES:
- A bracket atom stores an immutable virtual hydrogen count set by the writer.
- A bare atom stores a mutable virtual hydrogen count initially set to the smallest target valence. This count is decreased by the order of newly-created bonds. Should the virtual hydrogen supply become depleted, more can be obtained by graduating the atom to the next highest valence state.
In other words, a bracket atom defines a final virtual hydrogen count, whereas a bare atom defines an initial hydrogen count that will be updated as bonds are added.
For example, consider methanol (CO). On encountering the leftmost token (C), we create a carbon atom. The target valence for carbon is four, so we initialize the atom with four virtual hydrogens. The next atom (O) is created in the same way, except it is initialized with two virtual hydrogens as per the table. An elided single bond exists between the two atoms. To account for it, the virtual hydrogen counts of both atoms are decremented, leaving counts of 3 and 1, for the carbon and oxygen atoms, respectively. Had the bond been of order two (as in formaldehyde C=O), then two virtual hydrogens would have been deducted.
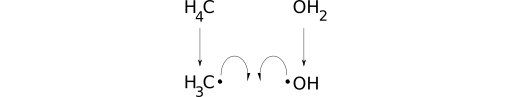
Now consider methylmagnesium chloride (C[Mg]Cl). First, a carbon atom with four virtual hydrogens is created. A magnesium bracket atom with zero hydrogens follows. When adding the elided single bond between these first two atoms, only the virtual hydrogen count of the first (C) is decremented. Next comes chlorine (Cl), which is assigned one virtual hydrogen from the table. To account for the elided bond between the last two atoms, only the virtual hydrogen count of chlorine (Cl) is decremented. Magnesium was created as a bracket atom, so its virtual hydrogen count will never change. The result is three, zero, and zero virtual hydrogens for carbon, magnesium, and chlorine, respectively.
This model has two obvious advantages. First, it brings implicit and virtual hydrogens closer together conceptually. Both bracket atoms and bare atoms have a virtual hydrogen count, it's just that the one on bracket atoms never changes in response to added bonds, whereas the one on bare atoms does. Second, this approach models molecular construction as electron flow, simplifying electron accounting. Bonding electrons can only flow from one bonding relationship (virtual hydrogens) to another (new bonds). Electrons can't be created or destroyed. The implications of this constraint will soon become apparent.
Aromaticity
The procedure for computing implicit hydrogen counts has so far left out the essential question of aromaticity. The meaning of this term has unfortunately been documented less than clearly over the years. For example, aromaticity in SMILES has been needlessly and confusingly associated with Hückel's Rule, an unwieldy concept at best. Beyond that, the use of a term with decades of prior history in organic chemistry is guaranteed to sow confusion.
In an effort to provide some clarity, I recently proposed a treatment of SMILES aromaticity that is consistent both internally and with existing use. The key concept is the π-subgraph. In a nutshell, a SMILES string specifies a molecular graph as a set of atoms and bonds between them. The π-subgraph is a subset of this molecular graph. Atoms are added to the π-subgraph by marking them as "aromatic" via lower-case element symbols (c, n, o, p, s, as, se). Both bare and bracket atoms eligible, but the labels "as" and "se" can only be used within brackets. Elided bonds between two aromatic atoms are marked as aromatic, as are bonds of type colon (:).
With this model of aromaticity in mind, we can approach the assignment of implicit hydrogen counts in SMILES with aromatic features.
Recall that implicit hydrogen count is defined as the difference between a target valence and an atom's encoded valence. Previously, valence was computed as a bond order sum. We now amend this definition to account for the π-subgraph.
We can think of the π-subgraph as a multi-atom bonding arrangement. Each atom in the π-subgraph contributes one electron, if available. Consistent with the model presented in the previous section, this electron comes from a hydrogen atom attached at atom's creation. If no implicit hydrogens are available, then none can be contributed.
Consider pyridine, represented by n1ccccc1. Using the convention that aromatic bonds yield a bond order of one, we obtain a valence of two for the nitrogen atom (1 + 1). The virtual hydrogen count is therefore one (3 - 2). Because the nitrogen atom participates in in the π-subgraph, its virtual hydrogen must be applied toward the π-subgraph. The result is a virtual hydrogen count of zero (3 - 2 - 1).
Next, consider the molecule represented by n1cccc1. It might look like pyrrole, but isn't. To understand why, perform the same analysis as with pyridine. One virtual hydrogen is present before accounting for the π-system. When considering the π-system, this virtual hydrogen is lost and its electron contributed to π-subgraph bonding. This leaves a virtual hydrogen count of zero (3 - 2 - 1) on the nitrogen atom.
Clearly this is incorrect. The nitrogen atom of pyrrole should have one hydrogen.
The solution is to take control of the hydrogen count for the nitrogen atom by wrapping it in brackets. Then, the correct virtual hydrogen count of one can be set: [nH]1cccc1.
Now consider furan, encoded as o1cccc1. Like pyrrole, the oxygen atom is bound to two neighbors through aromatic bonds. This atom's valence is therefore two (1 + 1). From the table, the target valence for the oxygen atom is also two. The hydrogen count of the oxygen atom, before considering the π-subgraph, is therefore zero (2 - 2). Because no virtual hydrogens are present, furan's oxygen atom can contribute no additional electrons. This phenomenon parallels that seen with "pruning" of certain atoms from the π-subgraph during kekulization.
Conclusion
SMILES supports two forms of hydrogen suppression: implicit hydrogen and virtual hydrogen. Virtual hydrogens apply to atoms within square brackets, which must either encode a hydrogen count or accept a default of zero. Bare atoms use implicit hydrogens; the hydrogen count for such atoms can only be determined by a valence model. The SMILES valence model is straightforward, but documentation has been less than clear. A comprehensive model, including corrections due to aromaticity, is presented here.