Stereochemistry and the V2000 Molfile Format
A previous article offered some reasons to adopt the V3000 molfile format. Although there are several, the one that gets the most attention is "enhanced stereochemistry" support. It should come as no surprise that the cost of this enhancement is increased complexity. Fortunately, V3000's stereochemistry model extends the one used in V2000. Unfortunately, the V2000 stereochemistry model is not exactly simple. This article explains V2000 stereochemistry in detail, both as a way to avoid data loss when working with the format, and as a way to understand what V3000 brings to the table.
Source Documentation
The following discussion is based on two documents published by the molfile format's corporate sponsor: CTFile Formats BIOVIA Databases 2020; and Chemical Representation 2018 SP1 ("the Guide"). The former covers syntax and the later covers semantics. I'll sometimes refer to both documents collectively as "the specification." The two documents must be read together and in their entirety to untangle the many nuances of the molfile format.
Defined and Undefined Stereocenters
Two concepts underpin the specification's treatment of stereochemistry: defined stereocenters and undefined stereocenters. The underlying concept, "stereocenter," was defined in 1983 by Mislow and Siegel as an atom whose "ligand permutation produces stereoisomers." For example, the tertiary carbon atom in alanine is a stereocenter because permutation of its ligands results in stereoisomers (specifically, enantiomers). The designations "defined" and "undefined" refer to whether or not a molfile notes the presence of a stereocenter.
A stereocenter is said to be "defined" if placed "at the narrow end" of an "Up or Down stereo bond" (Guide, p 9). Syntactically, a defined stereocenter is an atom connected to at least one bond whose "bond stereo" (sss) attribute is 1 ("Up") or 6 ("Down"), and whose "first atom number" (111) attribute equals the atom's identifier.
The Guide goes on to muddy the waters with one additional apparent restriction on an atom's "atom symbol" (aaa) property:
You can specify the stereoconfiguration at asymmetric tetrahedral centers (chirality centers) solely if the atom is one of the following: C, N, Si, P, As, S, Se or Te or any of their isoelectric equivalents (e.g. B-). [p 225]
This restriction occurs in the context of the section titled "Perception of Stereoconfiguration at Tetrahedral Centers." Similar language can be found in the 2011 version of the Guide:
You can specify the stereoconfiguration at asymmetric tetrahedral centers (chirality centers) at the following atom types: C, N, O, Si, P, or S. [p 24]
Here, atoms bearing both Se, Te appear to be disqualified as defined stereocenters, and there is no exception for isoelectronic equivalents. A previous article talked about other important changes to the molfile format that occurred after the 2011 revision.
Assuming that all conditions are met, a defined stereocenter implies that:
… something is known about the configuration at that center; either its absolute configuration, or its configuration relative to other stereogenic centers that are marked with stereo bonds. … [p 9]
In other words, a defined stereocenter presents readers with a choice: interpret the stereocenter as either an absolute or relative configuration.The next section explains a deciding factor.
If a stereocenter does not sit at the thin end of an Up or Down stereobond, it is considered undefined. An undefined stereocenter implies that:
… no information is known about the configuration of that stereogenic center. It could be either of two stereoisomers, or a mixture of the two. [p 9]
It's easy to miss the subtlety here. An undefined stereocenter represents one of three possibilities: an arbitrary configuration; the opposite configuration; or a mixture of both configurations. We'll return to this point later.
Atom Stereo Parity
The Guide talks at length about stereo parity, but this concept plays no role in V2000 molfiles. As noted by CTFile Formats, this atomic attribute (sss) is "ignored when read" (p 44). This lack of V2000 support for atom parity extends back to at least 2005. The only way to produce a defined stereocenter in V2000 files is to point the thin end of an Up or Down bond at it.
Chiral Flag
The interpretation of defined stereocenters is governed by the global chiral flag attribute. Syntactically, this attribute appears in a V2000 molfile's counts line (the fourth) from the 13th to 15th columns, inclusive. A value of 0 leaves the flag unset ("Off") and a value of 1 sets it ("On"). For example, a molfile with six atoms and five bonds whose chiral flag is set might have the following counts line:
6 5 0 0 1 0 0 0 0 0999 v2000
The purpose of the chiral flag is to convey to readers that the absolute configurations of all defined stereocenters is both known and defined as depicted. Setting the chiral flag has no effect on the interpretation of undefined stereocenters. The Guide uses the following language:
Setting the chiral flag to On indicates that the structure as drawn represents a single stereoisomer whose absolute stereochemistry is known. [p 225]
The Guide offers the example of (1R,2S)-(-)-trans-2-phenyl-1-cyclohexanol, encoded within a molfile whose chiral flag is set:
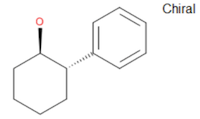
A molfile whose chiral flag is Off conveys to readers that no information is available on the absolute configuration at any defined stereocenter. They must only be interpreted as relative configurations. The Guide uses the following language:
Setting the chiral flag to Off indicates that the absolute configuration of the structure is unknown, but that the relative configurations of the stereogenic centers are known. The structure might represent either a single diastereomer or a mixture of the two stereoisomers.
The Guide goes on to consider the example of trans-2-phenyl-1-cyclohexanol, this time encoded within a molfile whose chiral flag is not set:
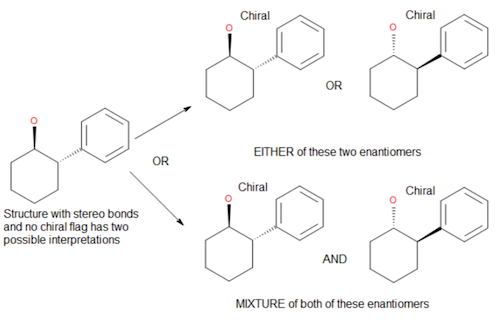
Because the chiral flag is Off, this molfile's defined stereocenters can only be interpreted in the relative sense. Futhermore, no information exists about the absolute configuration at either stereocenter. The molfile must therefore be interpreted in one of three possible ways: as one pure enantiomer; as the other pure enantiomer; or as a mixture of both enantiomers. The information needed to distinguish these possibilities does not exist.
The subtlety here bears repeating. The molfile very clearly does not encode a mixture of two enantiomers. Instead, it encodes three distinct possibilities: a single enantiomer; its antipode; or a mixture of the two.
Confusion on this point is not hard to find. A 2017 presentation on InChI, for example, makes the following statements that conflict with the Guide:
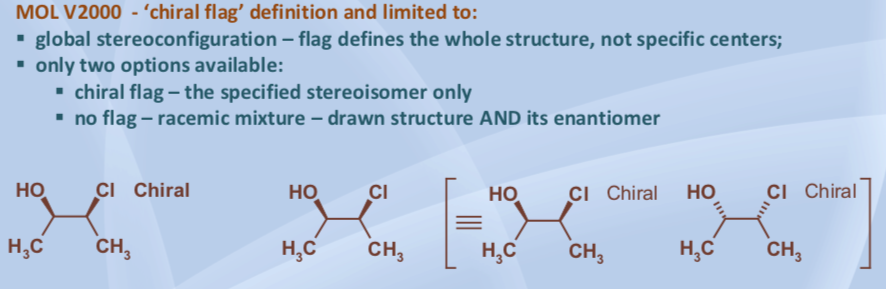
What about molfiles whose chiral flag is Off but which encode a single defined stereocenter? According to the Guide, such molfiles must be interpreted as encoding a mixture of enantiomers:
For structures with only one stereogenic center, a structure with stereo bonds but without the chiral flag represents a mixture of enantiomers [p 256]
Notice how this guidance contrasts with the way molfiles with two or more defined stereocenters are treated. Consider a molfile encoding alanine using one defined stereocenter, but with a chiral flag set to Off. This molfile must be interpreted as a mixture of enantiomers exclusively! The analogous trans-2-phenyl-1-cyclohexanol example was interpreted as one of three cases of which an enantiomeric mixture was one possibility, but the alanine example is a mixture only.
The following table summarizes the possible combinations of stereocenter type and chiral flag settings.
| Stereocenter | Chiral Flag | Configuration |
|---|---|---|
| Undefined | Off | either configuration OR both configurations |
| Undefined | On | either configuration OR both configurations |
| 1 Defined | Off | drawn configuration AND its inverse |
| 1 Defined | On | drawn configuration |
| > 1 Defined | Off | drawn configurations OR their collective inverse OR a mixture of both |
| > 1 Defined | On | drawn configurations |
The Guide does not discuss the case of a molfile having both defined and undefined stereocenters. The simplest interpretation would be that each type of stereocenter contributes independently. Consider a molfile whose chiral flag is Off and which encodes one defined stereocenter (A) and one undefined stereocenter (B). This molfile encodes the combinatorial result of applying the appropriate boolean logic in the table above. In other words, ONE of the following possibilities is encoded:
- AB + A'B
- AB' + A'B'
- AB + AB' + A'B + A'B'
Similar considerations would apply for a molfile whose chiral flag is Off and which encodes two defined stereocenters and one undefined stereocenter. And so on.
Mixture, not Racemate
The use of the term "mixture" is deliberate, both here and in the guide. The Guide uses the term in its traditional sense to denote the presence of two or more constituents. Notice, however, that the term implies nothing about the ratio of these constituents. Several sources incorrectly interpret the Guide's use of the term "mixture" to be equivalent to "racemate" or 50:50 mixture. This is not supported by my reading of the Guide, nor by the common use of the term "mixture."
Limitations
The Guide notes three main limitations with the V2000 stereochemistry model:
- There is no way to distinguish a stereochemical mixture from a single enantiomer in which all relative configurations are known.
- The chiral flag applies to the entire molfile. Even if multiple molecules (connected components) are present, they can not be treated differently.
- It's not possible to convey different levels of stereochemical uncertainty within the same molecule.
These specific limitations motivated the development of V3000's enhanced stereochemistry support.
Impedance Mismatch
The combination of defined/undefined stereocenters and chiral flag notation is somewhat unique to V2000 molfile. This idiosyncrasy can complicate the format's interconversion. For example, SMILES is a widely-used format devoid of the concept of chiral flag. What protocol should be used to translate a molfile containing multiple defined stereocenters, but with a chiral flag set to Off? Similar considerations apply when generating Standard InChIs, and many other formats.
This problem is compounded by the V2000 semantics around undefined stereocenters. Recall that these must be interpreted as one of three possibilities: pure isomer A; pure isomer B; or a mixture of both configurations. The SMILES documentation is silent on the meaning of stereocenters whose configuration is not defined. Although one organization might choose to use the same semantics, another organization might choose another interpretation. And things can get really dicey should a third format such as CDXML or CML become involved.
For some real-world examples of the trouble that can arise through simple conversions of SMILES and V2000 molfile, see:
It is likely that each organization facing this problem will arrive at its own solution. Although these solutions may be internally consistent in the short run, they can become internally inconsistent over the long run through turnover and loss of institutional memory. And it's extremely unlikely that two organizations will choose exactly the same solutions. The net effect will be data loss and confusion when organizations attempt to collaborate with each other, and even when an organization attempts to collaborate with its future self.
Conclusion
V2000 supports stereochemical configuration through two features: stereocenters and the chiral flag. Stereocenters are defined by pointing the narrow end of a stereobond at them. Otherwise, a stereocenter will be undefined. The global chiral flag attribute only modifies the interpretation of defined stereocenters, but in subtle and easily misinterpreted ways. These stereochemical idiosyncrasies are likely to cause data loss during interconversion of certain kinds of V2000 files with other formats, especially those involving vague semantics around undefined or partially-defined configurations.