Standard InChI
InChI is an IUPAC-sponsored project aimed at creating a distributed chemical identifier system. Centralized identifiers like CAS Registry Numbers operate through intermediaries who curate entries, resolve conflicts, and often charge a substantial fee for the service. InChI is different in that identifiers are generated deterministically through software, independent of an intermediary.
Useful though this distributed model may be, it's not without tradeoffs. The most important of these is the inherent tension between universality and flexibility. A universal identifier must necessarily be rigidly defined, but rigidity conflicts with chemistry's inherent messiness. This article explores the tension between these requirements within the context of a surprise I found during a benchmarking study.
About Standard InChI
InChI's approach to a universal yet flexible identifier system is standard InChI. Standard InChI is a kind of identifier generated through a specific configuration of the InChI software. Standard InChIs identify themselves with a prefix containing the letter S (e.g., InChI=1S/C2H6/c1-2/h1-2H3). Non-standard InChIs lack the S prefix (e.g., InChI=1/C2H6/c1-2/h1-2H3).
By default, all InChIs produced by the InChI software are standard. In other words, if no flags are passed, the software yields standard InChIs that can be compared to any other similarly-generated standard InChI. Several flags passed to the software will result in a non-standard InChI. The InChI User Guide notes that these flags include:
SUU. Always include omitted unknown/undefined stereo.SLUUD. Use different labels for omitted unknown/undefined stereo.RecMet. Preserve bonds to metal atoms.FixedH. Generate the fixed-hydrogens layer.KET, Account for experimental 1,3-tautomerism.15T. Account for experimental 1,5-tautomerism.SRel. Use relative stereo.SRac. Use racemic stereo.SUCF. Consume the molfile chiral flag.
A few flags can be passed without triggering non-standard output, including:
NEWPSOFF. Perceive stereocenters on both ends of a wedge/hash bond.DoNotAddH. Don't add hydrogen layer.SNon. Ignore all stereo information.LooseTSACheck. Enable detection of ambiguous (large-angled) bonds in cycles. Added in InChI v 1.06.
It should be emphasized that the distinction between standard and non-standard InChI only applies to properly-configured software, and to software operating directly on source molfiles. If anything comes between the molfile and the InChI software, all bets are off.
Trouble Brewing
As part of a long-standing effort to expand the range of computing environments on which InChI runs, I recently succeeded in compiling the InChI C source to WebAssembly. The result was InChI-Wasm, from which a build system, compiled Wasm file, and demonstration HTML document can be obtained.
Two kinds of performance were evaluated with a benchmark. In it, InChI-Wasm was matched with a counterpart library, also compiled from the InChI source, but as a native macOS dylib binary. Glue code in the form of JavaScript running on Node.js allowed a fair comparison of performance for the two ways of running InChI.
As a data set, I turned to something I hadn't used before: SureChEMBL. SureChEMBL is a patent database operated by the European Molecular Biology Laboratory (EMBL). Given that structures are drawn from the patent literature, the set seemed like a good performance test. Like many other public-facing chemical databases, SureChEMBL provides raw data files that can be downloaded in SDfile format and reprocessed. The presence of InChI field in the data set made it convenient to compare the identifiers reported by InChI-Wasm and SureChEMBL.
With the SureChEMBL data set and the two InChI versions in hand, two kinds of benchmarks were run. The first compared run time, finding that InChI-Wasm ran within a factor of two of the run time of the natively-compiled library.
The second performance test compared the output of InChI-Wasm with the InChIs reported by SureChEMBL. Out of 114,000 records, there were 155 discrepancies and 3 errors. Although this may seem low, it's not zero, which was my expectation. Due to time constraints, I simply noted these differences. But the question remained. Why do the InChIs reported by SureChEMBL not match those generated by InChI-Wasm?
Given the natively-compiled InChI library, it was easy to rule out compilation to WebAssembly as the source of the problem. Comparison of the output from both libraries (Wasm and native) yielded the same InChI output.
Wherever the discrepancy between InChI-Wasm and SureChEMBL identifiers may lie, it had nothing to do with WebAssembly.
Isolating the Problem
In many applications, InChI is run via the inchi-1 executable. This executable is compiled from C source that defines a main function accepting various inputs and which generates outputs.
But it's also possible to run InChI as a library by writing code that interacts with the InChI API. My benchmark used this method. A convenient entry point into the InChI library is the MakeInChIFromMolfileText function.
MakeINCHIFromMolfileText is a convenience function that accepts as input a molfile and flags (as discussed above), writing InChI output in response. The InChI API documentation has the following to say about MakeINCHIFromMolfileText:
This function creates InChI from Molfile supplied as a null-terminated string. That is, it automates reading/parsing Molfile, creation of InChI input and generation of InChI string. Notably, it relies on the same Molfile parser as inchi-1 executable thus ensuring that any correct caller will produce the same result as inchi-1. [emphasis added]
If for some reason the inchi-1 main method either didn't use MakeINCHIFromMolfileText or used it differently than documented, that could be the source of the SureChEMBL discrepancy.
To test this idea, I ran the inchi-1 executable on the decompressed SureChEMBL SDfile. To simplify comparison with the output from InChI-Wasm, I invoked the executable like so:
$ inchi-1 -AuxNone -OutErrInChI SureChEMBL_20210101_27.sdf
The resulting file (SureChEMBL_20210101_26.sdf.txt) contained two lines per entry: the first contained a record number and the second contained the InChI produced from the molfile. The -AuxNone flag suppresses auxiliary information, which is not needed. The OutErrInChI flag adds a line containing InChI=1S// in the event of a processing error. The final step was to remove the file header line, which was done either as a shell one-liner or with a text editor.
Next, I built a new test bench that uses InChI-Wasm to generate a text file with an identical format to the one created by the inchi-1 executable. The two files, one made by the official InChI executable and the other made by code using the InChI API and compiled to Wasm, could then be compared using the Unix diff utility.
The result showed both files to be identical. For completeness sake, I did the same analysis against inchi-1 compiled from both v 1.05 and v 1.06 (the most recent release and the one used by InChI-Wasm). The results were the same. The differences seen between InChIs reported by SureChEMBL and InChI-Wasm had nothing to do with WebAssembly, the version if InChI being used, or the way the InChI API was being accessed.
A Survey of Differences
To gain some insights into the 155 structures behind the identifiers that didn't match, I filtered the SureChEMBL SDfile file to retain only the conflicting records. Then I manually inspected several of them.
The most obvious thing to check was the LooseTSACheck flag. Recall that this flag was added in InChI v 1.06, and it enables the perception of stereochemistry on distorted tetrahedral centers within cycles. Setting this flag eliminated conflicts in about ten of the mismatched records.
Many of the remaining conflicts arose from structures containing metals. For example, SCHEMBL22832217 is the complex between methyl isobutyrate and potassium. What form of potassium, ion or atom, you ask? That's a good question.
The InChI produced by the InChI-Wasm benchmark was:
InChI=1S/C5H10O2.K.H/c1-4(2)5(6)7-3;;/h4H,1-3H3;;
Whereas the InChI reported by SureChEMBL was:
InChI=1S/C5H10O2.K/c1-4(2)5(6)7-3;/h4H,1-3H3;
The disagreement here revolves around the question of whether or not the potassium atom is bound to a hydrogen atom. This is no small question. The answer implies completely different molecular formulas and weights, and of course identifiers.
As noted on this blog previously, V2000 molfiles support virtual hydrogens through the awkward but workable mechanism of specifying valence. Implicit hydrogens are not assigned to metals, so setting valence on a metal is usually not productive in a molfile.
But InChI's approach to metals is a bit odd. As noted here, bonds to metals are always disconnected by default. However, virtual hydrogens are also assigned to metals! This leads to the counterintuitive situation in which a free metal will be saturated to satisfy InChI's own valence model, but those hydrogens won't be covalently attached.
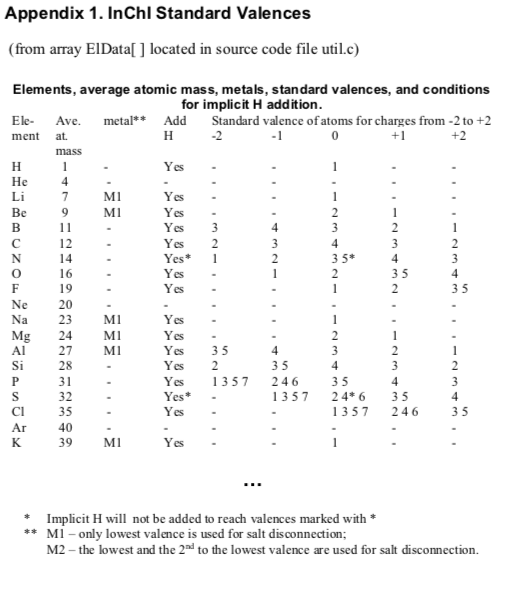
However, metal saturation can be disabled by setting the corresponding molfile valence flag to 15 (zero valence). In other words, InChI will saturate metals with hydrogens according to its own internal rules, but not if the metal's valence property is set by the molfile.
Taking all of this into account leads to the following hypothesis. At least in some cases, SureChEMBL's raw data file does not include the exact molfile used as input into the InChI software. Instead, something has processed the molfile in such a way as to set metal atom valences to zero.
This same pattern appeared to play out in other records that I spot checked. One or more transformations intervene between the reported molfile and InChI for some records. This could be a normalization utility that transforms valences, coordinates, and other properties to a second molfile which is them passed to InChI. Or it could be normalized data structures that get passed to a low-level InChI API call.
Additional classes of structure for which InChI-Wasm and SureChEMBL didn't agree included:
- several cases involving trivalent sulfur and ChemAxon V2000 valence extensions;
- many cases of stereochemistry-containing structures whose overlapping atomic coordinates are not readily apparent when viewing a 2D graphical representation;
- V3000 molfiles, which I did not check;
- as described previously, a few cases of
LooseTSACheckyielding different results while still yielding a standard InChI.
Why Consistency Matters
Reporting an inconsistent molfile/InChI pair, as appears to have been done by SureChEMBL, complicates the goal of reproducibility. Recall that InChI identifiers can't in general be parsed to yield molfiles. So it's reasonable to expect that when a database contains a molfile and an InChI, the molfile was directly used to generate the InChI. Confusion resulting from a mismatch of molfile and InChI is to be expected.
It seems likely that a molfile representing the actual SureChEMBL input to the InChI software exists somewhere. If so, why not report that instead of the raw input? Business rules could offer one explanation. If multiple sources submit molfiles into SureChEMBL, it may be reasonable to report them exclusively. The presence of both V2000 and V3000 molfiles in the data set lends support to the idea that representations are being obtained form multiple sources. Indeed, the README for the SureChEMBL download notes:
NB, this is the raw compound feed and is provided without further curation.
Conclusion
Standard InChIs make it possible to satisfy two constituencies: those who want a universal InChI identifier that can be used with any database, and those who want InChI to be flexible for special cases. But no matter how simple the idea may seem, nuances pop up in unexpected places. It's possible to set some flags, changing the identifier, but without triggering a non-standard InChI. It's also possible to pair a standard InChI with a molfile that didn't generate it. Both appear to have been done in the case of SureChEMBL.
Despite the issues I encountered with this particular data set, the identifiers obtained from InChI-Wasm and the official InChI distribution were identical. This bodes well for future use of InChI-Wasm as a platform-independent, high-performance alternative for natively-compiled InChI.