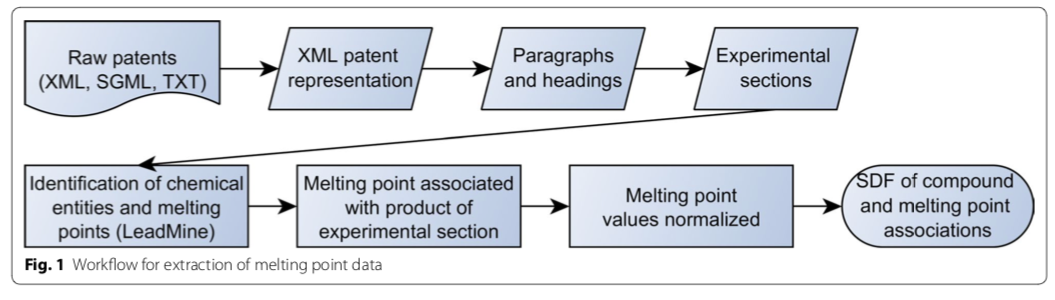
The NextMove Patent Reaction Dataset
Until recently, large databases of machine-readable chemical reactions were rare, constrained in their allowed uses, and extremely expensive. That changed in 2014 with the publication of a dataset of organic chemical reactions extracted from US patents and patent applications. What follows is an introduction to this dataset and a discussion of some of the ways it has been used.
Origins
The NextMove reaction dataset (NRD) is a collection of 3,305,795 reactions extracted from 9 million patent applications and issued patents from 1976-2016. It evolved out of work performed by Daniel Lowe for his PhD Dissertation at Cambridge. As part of a multi-student team whose work spanned several years, three key pieces of software were developed: OSCAR4, a suite of tools for processing free-form organic synthesis experimentals; ChemicalTagger, an automated annotation tool; and OPSIN, an IUPAC nomenclature parser. The latter has been the topic of multiple Depth-First articles, including this one. The NRD is CC-0 licensed, meaning that the creator waived any applicable copyright; there are no legal restrictions on redistribution or other uses.
Lowe's dissertation work was improved by NextMove Software to create NRD. In 2014 the publication of the complete dataset was announced. Around the same time, NextMove described a "gold standard" human-annotated patent corpus that it had created. The availability of both datasets should provide a sound foundation for those wanting to improve on existing extraction tools and develop new approaches.
The NRD now stands as the largest, machine-readable, fully-reusable collection of organic reactions in existence.
Downloading the Dataset
The full dataset can be downloaded from Figshare.
Dataset Contents
The download from Figshare consists of five files:
2001_Sep2016_USPTOapplications_cml.7zAnnoted patent applications from 2001-2016 in XML format, grouped by issuing year into directories.1976_Sep2016_USPTOgrants_cml.7zAnnotated patent grants from 1976-2016 in XML format, grouped by issuing year into directories.1976_Sep2016_USPTOgrants_smiles.7zReaction SMILES for patent grants from 1976-2016 as a single Reaction SMILES file (.*rsmi).2001_Sep2016_USPTOapplications_smiles.7zReaction SMILES for patent applications from 2001-2016 as a single Reaction SMILES file (.*rsmi).xml_xsd.zipSchema Definition for XML files.
Reaction SMILES (*.rsmi) Files
The Reaction SMILES (*.rsmi) files contain the following fields, in order of appearance:
- reaction SMILES;
- patent number;
- paragraph number;
- year;
- text-mined yield;
- calculated yield.
A few words on the difference between "text-mined" yield and "calculated" yield are in order. "Text-mined" yield refers to the yield, expressed as a percentage, reported in the experimental section. "Calculated" yield refers to the yield calculated from the quantities of reagents used and product obtained.
For example, US20010000038A1 paragraph 0256 reports a yield of 86%. However, based on the molecular weights of the limiting reagent (120.185 g·mol-1) and product (224.717 g·mol-1), a yield of 86.9% was actually obtained. In other words, the procedure in this patent application slightly under-reports its experimental yield. This discrepancy is captured in the files.
Chemical Markup Language (*.cml) Files
The dataset's CML (*.cml) files contain the following major headings:
- Source. Bibliographical information including paragraph and patent (application) ID.
- Text. The complete experimental text.
- Reaction SMILES. Line notation describing the molecular structure of reactants, products, and reagents (see below).
- Reactant list. Enumeration of reactants, including information such as name, SMILES, and InChI.
- Product list. Enumeration of products, including such information as name, reported and computed yields, SMILES, InChI, appearance, and state
- Spectator list. Includes such components as solvents and quantities used.
- Reaction action list. An ordered list of steps to perform the synthesis. Each step includes: the full text being represented; the action being performed (e.g., "add"); the components being acted on; the conditions (e.g., nitrogen atmosphere); the time; and temperature.
Viewing Reaction SMILES
Reactions are encoded as Reaction SMILES. In this line notation, reactants and products are expressed using SMILES notation, and are separated by the greater than symbol (>). For example, the first record in the 2001_Sep2016_USPTOapplications_cml.7z data set contains this Reaction SMILES:
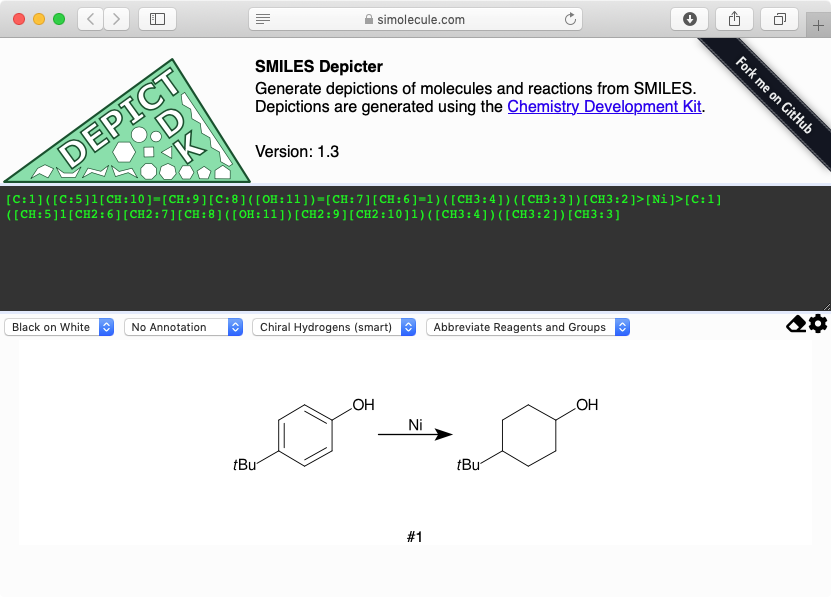
[Cl-].[Al+3].[Cl-].[Cl-].[Cl:5][CH2:6][CH2:7][CH2:8][C:9](Cl)=[O:10].[C:12]1([CH:18]([CH3:20])[CH3:19])[CH:17]=[CH:16][CH:15]=[CH:14][CH:13]=1>C(Cl)Cl>[Cl:5][CH2:6][CH2:7][CH2:8][C:9]([C:15]1[CH:16]=[CH:17][C:12]([CH:18]([CH3:20])[CH3:19])=[CH:13][CH:14]=1)=[O:10]
Reaction SMILES supports atom mapping through SMILES classes (the numbers appearing after the colon for bracketed atoms). See below for a discussion of reaction mapping.
The following image represents this reaction:
This image brings up two important points. First, molecular entities to the left, right, and above the reaction arrow are captured. Second, the metal appears in its disconnected, ionic form, despite the fact that most chemists would consider the Al-Cl bonds in aluminum chloride to be covalent. This latter point was a common feature of the records I spot-checked.
The latter point results from behavior in pre-2017 releases of OPSIN. Specifically, aluminum-halogen bonds were considered to be ionic based on a simplistic electronegativity calculation. OPSIN now considers Al-X bonds, where X is a halogen other than fluorine, to be covalent. Should the distinction be important, the NRD appends extended SMILES grouping notation to relevant Reaction SMILES.
A convenient tool for visualizing Reaction SMILES is CDKDepict. Clear the default text, then entire the Reaction SMILES as-is. Note that the XML files escape the greater than symbol (">"), and that it will need to be unescaped for use with CDKDepict. The service exposes URLs that can be used to later reference a Reaction SMIlES. See for example, this one.
Reaction Mapping
All reactions in the dataset have been mapped. Reaction mapping is a process whereby product atoms are mapped to the their corresponding reactant and/or reagent atoms. The NRD includes mappings from the reaction mapper functionality built into the Indigo Toolkit. A few other reaction mappers are currently available. Custom mappings from one of them may be more appropriate than the NRD defaults.
Applications
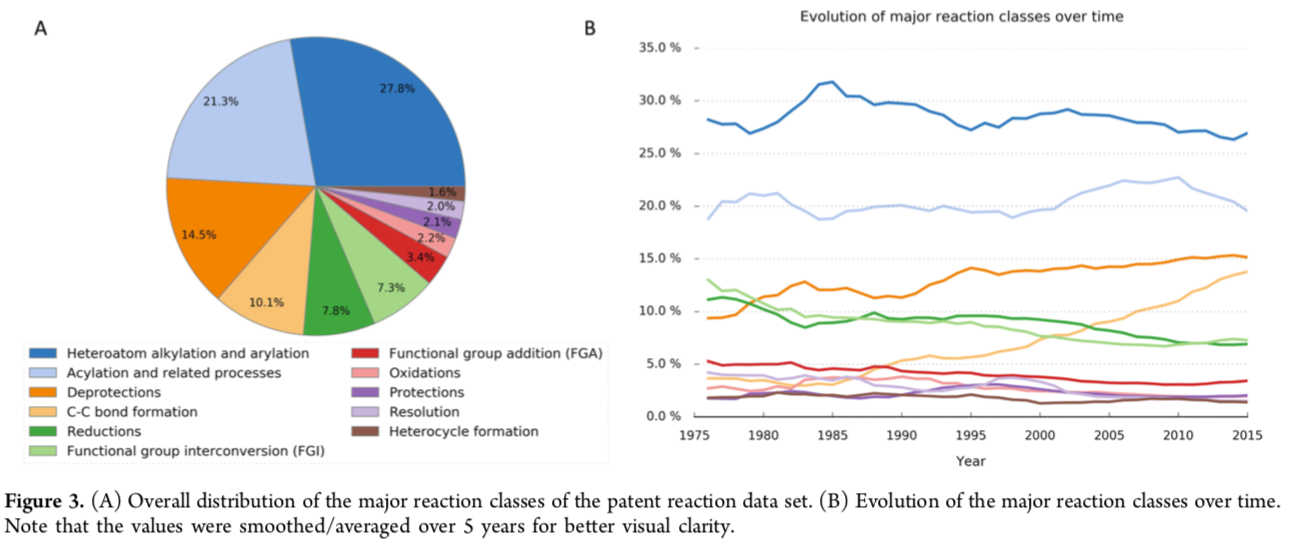
A corpus as comprehensive and practical as the NextMove dataset can clearly be used in many ways. One of the first applications examined the characteristics of reactions and synthetic targets for medicinal chemistry over time.
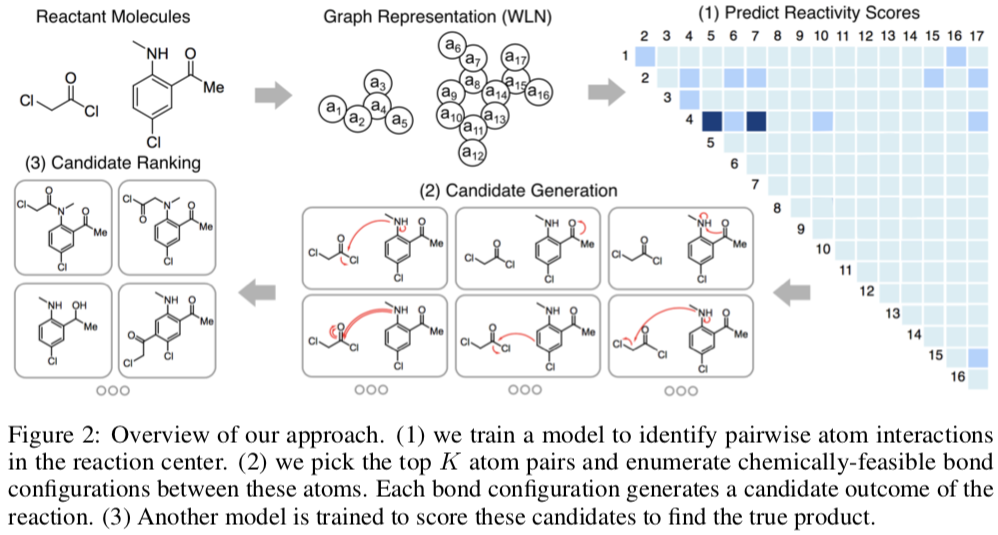
The NRD has been used in several projects aimed at developing machine learning models. For example, a group at MIT used a 480K-reaction subset of the the NextMove dataset to build a template-free reaction product prediction system.
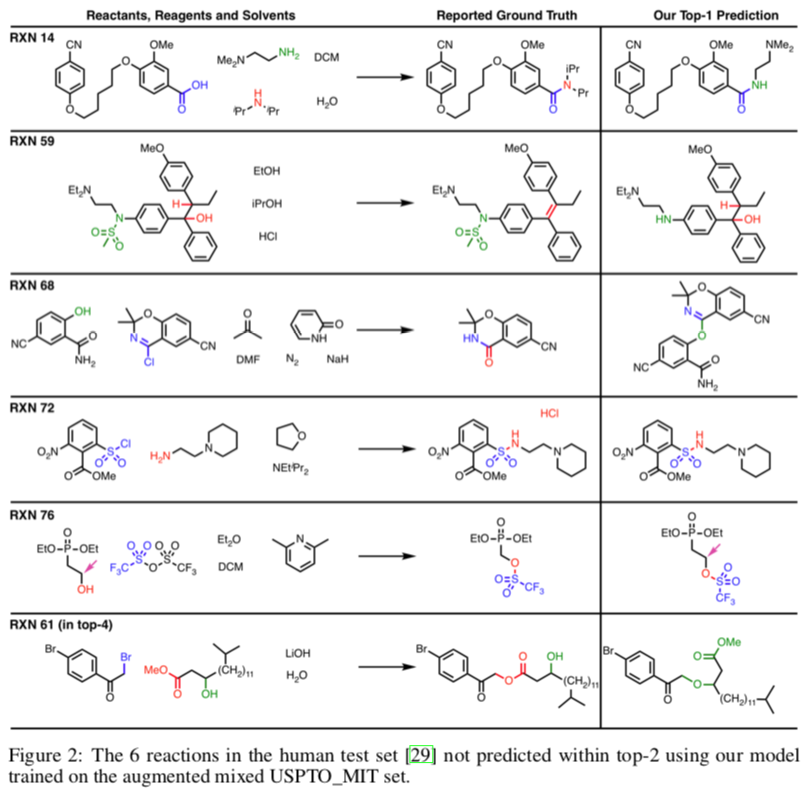
A similar approach was used by a group at IBM to develop a reaction outcome predictor. Among other useful features, it could estimate its own accuracy, which was determined to be 89%. An implementation is available online.
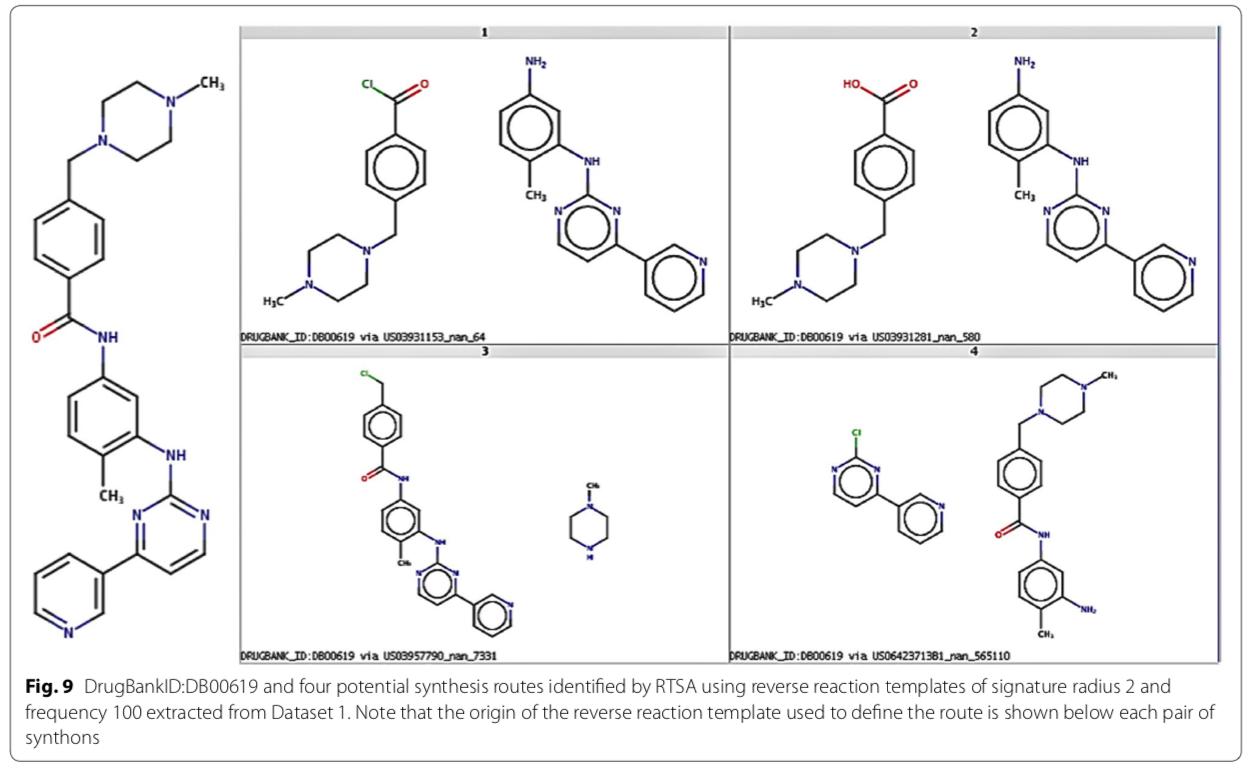
Recently, a group from Lilly described a data-driven retrosynthetic analysis engine trained with the NRD. The implementation is available on GitHub.
An approach similar to the one used to generate NRD was used to harvest a large collection of experimentally-determined melting points for use in building a melting point model.
Conclusion
Organic synthesis underpins much of modern society. As such, it's hard to overstate the utility of an open, machine-readable, freely-reusable, annotated reaction corpus as large and complete as the NextMove reaction dataset. The theoretical and practical applications that have already appeared hint at some of the things that are now possible.