Cheminformatics Data Pipelining with KNIME - Getting Started
Whether you consider visual workflow/data pipelining environments like Pipeline Pilot useful or not, there's no denying they're popular. Within the last few years, KNIME has joined Pipeline Pilot as a widely-used visual programming platform. But unlike Pipeline Pilot, KNIME is released under an open source license, which makes it interesting from the perspective of disruptive technology. This article, the first in a series, examines KNIME as a tool for cheminformatics data pipelining.
KNIME for Cheminformatics
In a blog post earlier this year, Andrew Dalke presented a summary of his experiences with KNIME. Considering that visual programming environments like KNIME are more accessible to "non-programmers" and from this a significant amount of their attraction derives, Dalke attempted to do some simple things with KNIME - and pretty much failed:
As far as I can tell, there's no way with the default nodes to do much of anything with KNIME. I assume there are additional packages which I can install, but why aren't there more useful CDK nodes as part of the standard installation? …
First the bad news: most of what Andrew previously reported appears to still true. The default KNIME installation for OS X does offer some components for cheminforamtics, but the selection is rather small. More frustrating, though, is the general lack of information on how to do something simple with KNIME in cheminformatics - and succeed. That's in part the problem this series attempts to address.
A Simple Workflow: Lipinski Rule of Five
I started with a simple goal: read a one-record SD file, calculate Lipinski Rule of 5 violations, and write the output back as an SD file containing a new Lipinski Rule of Five field. After some stumbling around… success!
Here's my workflow:
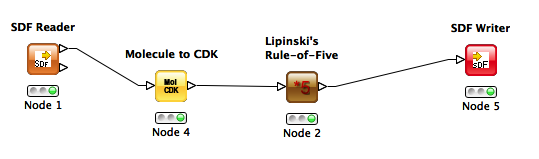
Gotchas
One of the problems you'll almost certainly hit when using KNIME for cheminformatics is the following error message:
"No CDK compatible column in input table"
This is the same error Andrew found, and as his post describes, the KNIME support forum recognizes this as a known issue:
We obviously need to improve on the error messages. You need to process the output of the SD reader with the "Molecule to CDK" node, which will parse the structures into an appropriate format for the Lipinski node. Reason is that the Lipinski node is contributed from the CDK plugin, so it needs its desired input format.
I have no idea what this means. If I had to guess, I'd say that the raw output from the SD Reader is the text of the molfile itself - but the Lipinski node is expecting an in-memory CDK Molecule object. The converter does that job. Maybe it would be far better for components to be intelligent about the format they need and if they don't have it, generate it themselves.
One more gotcha: the "Molecule to CDK" node is not especially easy to find. It's located under the "Translators" tab, and comes with a counterpart, "CDK to Molecule", whose function is a bit of a mystery at this point because I could write my SD file without problem after converting structures to CDK.
Still another gotcha: I don't think the Linux or Windows distributions come pre-packaged with any cheminformatics nodes - at least there were none I could find. The CDK cheminformatics nodes seem to be unique to the Mac distribution (v2.2.2). YMMV.
Conclusions
After a shaky start, we've managed to set up and run a simple cheminformatics workflow in KNIME. What else can we do with with cheminformatics data pipelining in KNIME? Future articles will describe some possibilities.