Simple 3D Conformer Generation with Smi23D
Three-dimensional conformer generation is a common problem in cheminformatics. The most convenient and generally-useful method for creating chemical structures is the 2D chemical structure editor; applications that require three-dimensional representations need a way to generate reasonable coordinates from 2D user input. Until recently, there were no options for doing so with Open Source software. This article shows how the Open Source package smi23d can be used to convert ordinary SMILES strings into three-dimensional molfile representations.
About smi23d
smi23d uses a two-stage process to generate 3D coordinates.; an initial pass with smi2sdf generates rough coordinates and subsequent refinement by mengine results in the final coordinates. The package was originally written in C by Kevin Gilbert and updated by Rajarshi Guha. As part of what appears to be a growing trend in cheminformatics, smi23d is licensed under the highly-permissive Apache License.
On a related note, the source code for a program called Frog is reportedly on its way into the Open Babel project.
Prerequisites
To build smi23d, you'll need to install Scons, a Make-like build utility written in Python. I was able to install the Scons rpm on my Linux system without a problem. smi23d uses no other dependencies.
Download smi23d
smi23d can be downloaded with Subversion:
svn co https://cicc-grid.svn.sourceforge.net/svnroot/cicc-grid/cicc-grid/smi23d/trunk smi23d
Building smi23d
With the source code in place, compilation is just a matter of running Scons:
cd smi23d
scons
...
Once the sources are compiled, we'll want to configure our system a bit:
cd build
ls
mmff94.prm mmxconst.prm
cp ../src/smi2sdf/smi2sdf .
cp ../src/mengine/mengine .
The two files mmff94.prm and mmxconst.prm are parameter files needed by both smi2sdf and mengine.
With smi2sdf and mengine both in the build directory, we can create a simple test with the SMILES for Ivabradine:
vi test.smi
...
$ cat test.smi
CN(CCCN1CCC2=CC(=C(C=C2CC1=O)OC)OC)C[C@H]3CC4=CC(=C(C=C34)OC)OC
With everything ready to go, we can begin Stage one:
./smi2sdf test.smi
Found 1 structures in test.smi
field : MMX
Atom Types: 169
Bonds: 580 Bond3: 0 Bond4: 0 Bond5: 0
Angle: 434 Angle3: 41 Angle4: 60 Angle5: 0
Torsion: 697 Torsion4: 58 Torsion5: 0
Vdw: 172 OOP: 91 Dipole: 474 Charge: 0 Improper: 0
STBN: 26 ANGANG: 0 STRTOR: 0 VDWPR: 4
Input file = test.smi
Output file = output.sdf
Param file = mmxconst.prm
Log file = error.log
Inorganic file = test_inorg.smi
Structure: 0 CN(CCCN1CCC2=CC(=C(C=C2CC1=O)OC)OC)C[C@H]3CC4=CC(=C(C=C34)OC)OC
You can view the result in an application like Jmol:
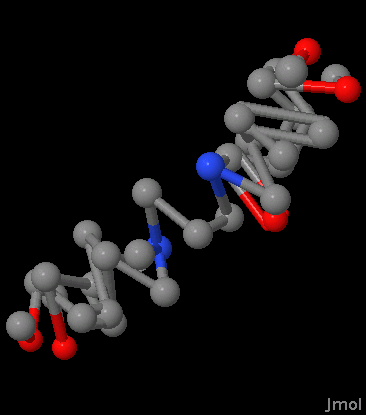
It's not much to look at, but we're not quite done yet.
Stage two is accomplished by using the output of Stage one as input to mengine:
./mengine -o optimized.sdf output.sdf
field : MMX
Atom Types: 169
Bonds: 580 Bond3: 0 Bond4: 0 Bond5: 0
Angle: 434 Angle3: 41 Angle4: 60 Angle5: 0
Torsion: 697 Torsion4: 58 Torsion5: 0
Vdw: 172 OOP: 91 Dipole: 474 Charge: 0 Improper: 0
STBN: 26 ANGANG: 0 STRTOR: 0 VDWPR: 4
field : MMFF94
Atom Types: 181
Bonds: 448 Bond3: 0 Bond4: 0 Bond5: 0
Angle: 1801 Angle3: 21 Angle4: 61 Angle5: 0
Torsion: 674 Torsion4: 38 Torsion5: 95
Vdw: 182 OOP: 112 Dipole: 0 Charge: 0 Improper: 0
STBN: 286 ANGANG: 0 STRTOR: 0 VDWPR: 0
We now have a file called output.sdf. As you can see, it's a pretty good 3D representation of Ivabradine:
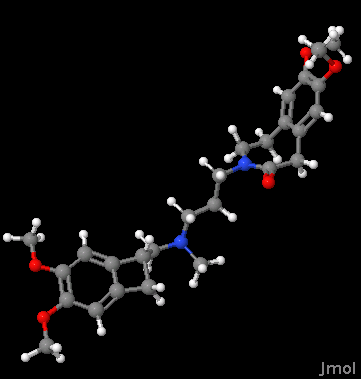
Conclusions
In this tutorial, we've seen how the Open Source program smi23d can be used to assign reasonable 3D coordinates to an arbitrary SMILES string. One very practical use of smi23d would be to process the output of 2D chemical structure editors prior to use in a 3D program. Future articles will discuss some of the possibilities.