Hacking PubChem - Visually Inspect Results for CAS Number and Keyword Searches
A recent article described how PubChem could be used to quickly search for CAS numbers. Although useful, the approach is limited in that only an array of PubChem CIDs was returned. What would be really useful would be a simple way to create a report with entries hyperlinked into the PubChem site itself to aid in visual inspection. In this tutorial, we'll see how an HTML template and a few extra lines of code can do just that.
The Template
Ruby supports a number of HTML templating mechanisms. In this example, we'll use an ERB template resurrected from the Molbank graphical table of contents tutorial:
<html>
<head>
<title>
<%= "PubChem Search for #{term}" %>
</title>
</head>
<body>
<h1>
<%= "Search: #{term}" %>
</h1>
<table>
<tr>
<% col = 0 %>
<% cids.each do |cid| %>
<td>
<% image = "http://pubchem.ncbi.nlm.nih.gov/image/imgsrv.fcgi?cid=#{cid}" %>
<% summary = "http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=#{cid}" %>
<a href="<%= summary %>">
<img src="<%= image %>" border="2"></img>
</a>
<center>
<span style="font-size: 8px">
<a href="<%= summary %>"><%= "CID-#{cid}" %></a>
</span>
</center>
</td>
<% col += 1 %>
<% if col > 5 %>
<% col = 0 %>
</tr>
<tr>
<% end %>
<%end %>
</tr>
</table>
</body>
</html>
The above template uses a search term and an array of CIDs to build a table of results. Each cell in the table contains a color 2D image and the CID, both hyperlinked into PubChem itself.
Saving this library to a file called template.rhtml is all we need to do.
The Library
The library is a modification of the one shown in the previous article in this series:
require 'rubygems'
require 'mechanize'
require 'erb'
module PubChemTerms
def report term
cids = get_cids term
erb = ERB.new(IO.read("template.rhtml"))
File.open "output.html", 'w+' do |file|
file << erb.result(binding)
end
end
def get_cids term
agent = WWW::Mechanize.new
page = agent.get "http://www.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pccompound&retmax=100&term=#{term}"
(page.parser/"id").collect {|id| id.innerHTML}
end
end
The method report accepts a search term and uses our template to render a report.
Testing
By saving the above library in a file called pubchem.rb, we can search by keyword via interactive ruby (irb):
irb
irb(main):001:0> require 'pubchem'
=> true
irb(main):002:0> include PubChemTerms
=> Object
irb(main):003:0> report 'esomeprazole'
=> #<File:output.html (closed)>
This produces a file called output.html that can be viewed with any browser:
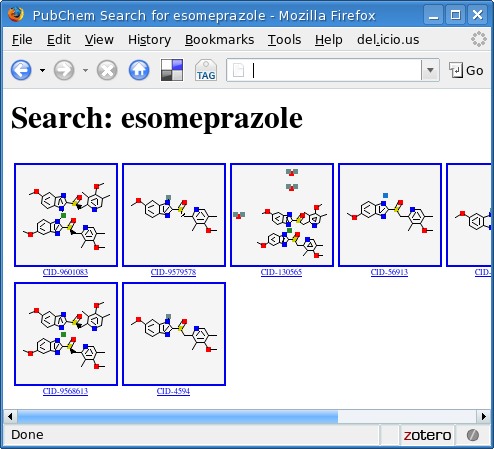
As in the original version of the library, we can also query by CAS number:
irb
irb(main):001:0> require 'pubchem'
=> true
irb(main):002:0> include PubChemTerms
=> Object
irb(main):003:0> report '119141-88-7'
=> #<File:output.html (closed)>
Conclusions
The simple approach outlined here could be extended in many ways. For example, we could easily retrieve molfiles based on keyword or CAS number search. We could pipe queries together or work with query lists. We could blend in ChemSpider data. We could even build a simple Web application (with Rails) that returned customized reports. Mixing in Ruby CDK or Ruby Open Babel offers still more possibilities.
Increasingly, the most important question in cheminformatics is not "What can we build?", but rather "What should we build?" Success in this new world requires a much deeper understanding of how cheminformatics software is being used by real chemists and where it's not.