A Molecular Language for Modern Chemistry: FlexMol and Axial Chirality
A recent article introduced FlexMol as a molecular language with the unique capability of encoding axial chirality. A previous article showed how E/Z geometrical isomerism is encoded with FlexMol. Using the popular chiral reagent and ligand 1,1'-bi-2-naphthol (BINOL) as an example, this tutorial will illustrate in detail how axial chirality is encoded in FlexMol.
Configuration or Conformation?
In contrast to configurational stereoisomers, conformational stereoisomers can be interconverted through bond rotations. So we'll need to use a conformationWheel to represent stereochemistry in BINOL - just as we did with 2-butene. For more rigorous definitions of these concepts, see the original specification by Dietz.
(R)-BINOL
A FlexMol representation and associated atom numbering scheme (R)-BINOL are show below:
<!-- (R)-BINOL -->
<?xml version="1.0" standalone="yes"?>
<molecule>
<constitution>
<atoms>
<atom id="C0" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C1" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C2" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C3" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C4" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C5" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C6" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C7" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C8" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C9" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C10" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C11" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C12" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C13" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C14" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C15" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C16" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C17" symbol="C" hydrogens="1" ionization="4"></atom>
<atom id="C18" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="C19" symbol="C" hydrogens="0" ionization="4"></atom>
<atom id="O20" symbol="O" hydrogens="1" ionization="2"></atom>
<atom id="O22" symbol="O" hydrogens="1" ionization="2"></atom>
</atoms>
<bonding>
<bond source="C0" target="C1" bondingElectrons="2"></bond>
<bond source="C1" target="C2" bondingElectrons="2"></bond>
<bond source="C2" target="C3" bondingElectrons="2"></bond>
<bond source="C3" target="C4" bondingElectrons="2"></bond>
<bond source="C4" target="C5" bondingElectrons="2"></bond>
<bond source="C0" target="C5" bondingElectrons="2"></bond>
<bond source="C0" target="C6" bondingElectrons="2"></bond>
<bond source="C6" target="C7" bondingElectrons="2"></bond>
<bond source="C7" target="C8" bondingElectrons="2"></bond>
<bond source="C8" target="C9" bondingElectrons="2"></bond>
<bond source="C9" target="C1" bondingElectrons="2"></bond>
<bondingSystem bondingElectrons="10">
<connections>
<atomPair source="C0" target="C1"></atomPair>
<atomPair source="C1" target="C2"></atomPair>
<atomPair source="C2" target="C3"></atomPair>
<atomPair source="C3" target="C4"></atomPair>
<atomPair source="C4" target="C5"></atomPair>
<atomPair source="C0" target="C5"></atomPair>
<atomPair source="C0" target="C6"></atomPair>
<atomPair source="C6" target="C7"></atomPair>
<atomPair source="C7" target="C8"></atomPair>
<atomPair source="C8" target="C9"></atomPair>
<atomPair source="C9" target="C1"></atomPair>
</connections>
</bondingSystem>
<bond source="C10" target="C11" bondingElectrons="2"></bond>
<bond source="C11" target="C12" bondingElectrons="2"></bond>
<bond source="C12" target="C13" bondingElectrons="2"></bond>
<bond source="C13" target="C14" bondingElectrons="2"></bond>
<bond source="C14" target="C15" bondingElectrons="2"></bond>
<bond source="C10" target="C15" bondingElectrons="2"></bond>
<bond source="C10" target="C16" bondingElectrons="2"></bond>
<bond source="C16" target="C17" bondingElectrons="2"></bond>
<bond source="C17" target="C18" bondingElectrons="2"></bond>
<bond source="C18" target="C19" bondingElectrons="2"></bond>
<bond source="C19" target="C11" bondingElectrons="2"></bond>
<bondingSystem bondingElectrons="10">
<connections>
<atomPair source="C10" target="C11"></atomPair>
<atomPair source="C11" target="C12"></atomPair>
<atomPair source="C12" target="C13"></atomPair>
<atomPair source="C13" target="C14"></atomPair>
<atomPair source="C14" target="C15"></atomPair>
<atomPair source="C10" target="C15"></atomPair>
<atomPair source="C10" target="C16"></atomPair>
<atomPair source="C16" target="C17"></atomPair>
<atomPair source="C17" target="C18"></atomPair>
<atomPair source="C18" target="C19"></atomPair>
<atomPair source="C19" target="C11"></atomPair>
</connections>
</bondingSystem>
<bond source="C9" target="C19" bondingElectrons="2"></bond>
<bond source="C8" target="O20" bondingElectron="2"></bond>
<bond source="C18" target="O21" bondingElectron="2"></bond>
</bonding>
</constitution>
<conformation>
<conformationWheel>
<gammaSequence source="C19" target="C9">
<connections>
<atomPair source="C9" target="C19"></atomPair>
</connections>
</gammaSequence>
<halfPlane>
<lower atom="C11"></lower>
</halfPlane>
<halfPlane>
<upper atom="C1"></upper>
</halfPlane>
<halfPlane>
<lower atom="C18"></lower>
</halfPlane>
<halfPlane>
<upper atom="C8"></upper>
</halfPlane>
</conformationWheel>
</conformation>
</molecule>
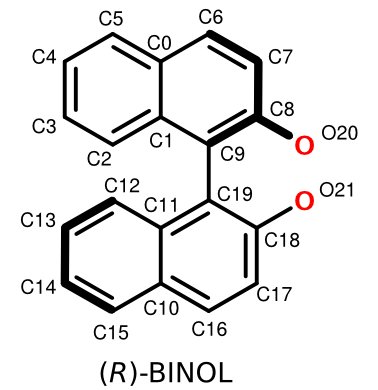
We've elected to represent BINOL's two pi-systems as ten-atom, ten-electron bondingSystems. We could have just as easily represented each naphthalene ring using alternating single/double bonds containing two and four electrons, respectively. For an explanation of multi-atom pi-system bonding in FlexMol, see this article.
The stereochemically-relevant part of this document is contained within the conformation element. A gammaSequence, or conformational axis, is defined along with four non-empty halfPlanes. Notice how the basic structure of this conformation element closely resembles the one for 2-butene.
To better visualize the the conformation element of (R)-BINOL, consider the following diagram:
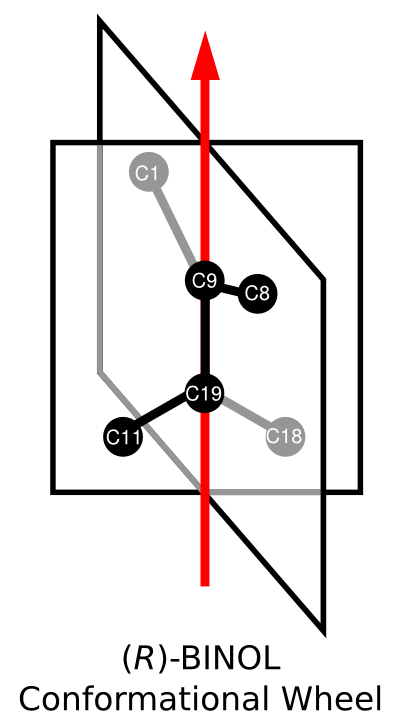
The conformationWheel defines a conformational axis vector from atom C19 to atom C9. Arranged about this axis in a clockwise fashion are four non-empty halfPlanes. Picking an arbitrary halfPlane to start with, atom C11 is positioned first in the lower half. This is then followed by the next halfPlane, which contains atom C1 in its upper half. The next halfPlane contains atom C18 in the lower half. Finally, atom C8 is located in the last halfPlane's upper half.
This procedure completely specifies the axial chirality of (R)-BINOL. Notice how no arbitrary stereodescriptors or chiral templates were used. Of course, we could derive the Cahn-Ingold-Prelog stereodescriptor of (R), given the right software.
Many representations of the same chiral axis are possible, just as each connection table can be represented in many different ways. For example, we could have started the conformation element with the halfPlane containing atom C1. In this case, the ordering of atoms would be C1, C18, C8, C11. Similarly, the orientation of our chiral axis could have been defined from atom C9 to atom C19. In this case the ordering of halfPlanes would be reversed, and the upper/lower designations would be inverted.
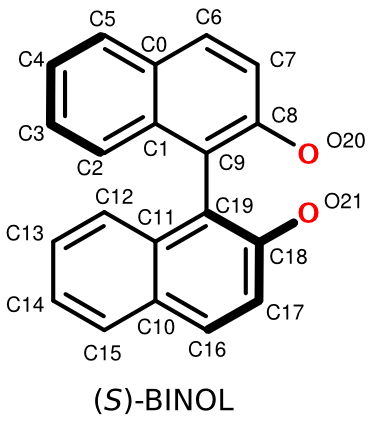
(S)-BINOL
How is (S)-BINOL encoded in FlexMol? As you might expect, completely analogously to the (R) enantiomer:
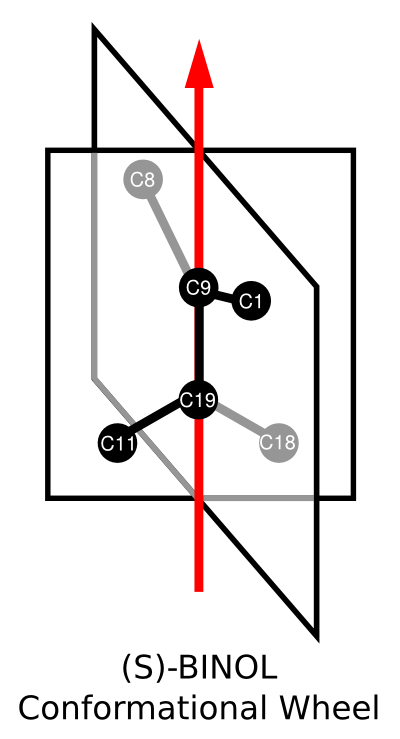
As with (R)-BINOL, we can create a diagram representing the conformationWheel of (S)-BINOL:
Conclusions
FlexMol completely encodes axial chirality using just a few basic XML elements, rather than chiral templates or stereodescriptors. These were, in fact, the same elements used to encode alkene geometrical isomerism. This modular approach to stereoisomerism results in an extensible system. Future articles will discuss other forms of stereoisomerism that can be represented in FlexMol, including the all-important tetrahedral stereogenic center.