Anatomy of a Cheminformatics Web Application: InChIMatic
InChI is an open molecular identifier system. Although InChIs obviate the need for a central registration authority, they are complex enough that they must be generated by computer. Currently, a few desktop molecular editors can generate InChI identifiers. But wouldn't it be more convenient if this capability existed in a simple Web application that could be used from any computer - anywhere? This article describes a Web application called "InChIMatic", which does just that.
In this article, I'll show how Java Molecular Editor (JME), a lightweight 2-D structure editor, can be extended to produce InChI identifiers through server-side software written in Ruby, rather than by extending the applet with Java code.
Downloads and Prerequisites
InChIMatic requires Ruby on Rails and the Rino InChI toolkit. Both of these libraries can be installed using the RubyGems packaging system.
The complete InChIMatic source package can be downloaded from RubyForge. For convenience, a copy of JME is included with the distribution. The author, Peter Ertl, has kindly given permission for the bundled JME applet to be used with InChIMatic. For other uses, consult the JME homepage.
Running InChIMatic
cd inchimatic-0.0.2
ruby script/server
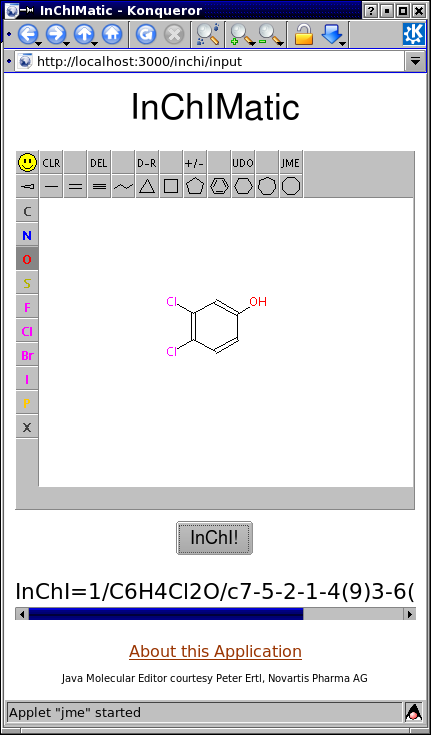
Pointing your browser to http://localhost:3000/inchi/input, drawing a structure in the JME window, and pressing the "InChI!" button will produce the corresponding InChI in the area below.
Behind the Scenes
The JME applet itself provides no capabilities for generating InChI identifiers. This functionality is instead provided by the Rails server via the Rino InChI library.
Let's say Susan wants to get the InChI for 3,4-dichlorophenol. After entering the structure into the JME window, she presses the "InChI!" button. This sets in motion the following sequence of events:
- The JavaScript function
writeMolfileis called. This retrieves a molfile representation of 3,4-dichlorophenol from JME, which is then written to to the hidden fieldmolfile. - A Rails listener notices that the hidden text field has been updated and so invokes the InChIMatic
ajax_inchiaction. This is a Rails Ajax action that will update only a portion of the InChIMatic window. For more detail on this Rails Ajax technique, see the previous Anatomy of a Cheminformatics Web Application article. - The
ajax_inchiaction retrieves the contents of the hiddenmolfilefield. This molfile is then used to generate an InChI using Rino. This InChI is then saved to the instance variableinchi. - The contents of the InChIMatic area partitioned by the
resultsdivare then updated with the InChI obtained in Step 3. The JME applet itself is unaffected by this operation, allowing Susan to further elaborate her molecule, if she'd like.
So What? Re-Thinking the Role of Applets
JME is, by itself, incapable of generating InChIs. Yet InChIMatic provides this capability as if it existed natively. In other words, a lightweight, fast-loading, and responsive 2-D editor can be extended on the server side, rather than on the client side. The difference is imperceptible to the user, but ripe with potential for the developer.
One of the most common, and completely valid, complaints about Java applets is that they take too long to load. Offloading some of the functionality currently being bundled in applets onto a Web server offers one way to combat the problem. Furthermore, combining Java applets with Ajax and powerful Web application frameworks like Ruby on Rails offers virtually limitless opportunities to re-think the role of Java applets in Web application development.
Conclusions
JME's strength comes, perhaps ironically, from its limited functionality. By using some simple Web programming techniques, JME can be extended with server-side programming. The advantages, compared to extending the JME applet itself with Java on the client side, are significant. Future articles in this series will explore some of the possibilities.