Agile Chemical Informatics Development with CDK and Ruby: RCDK-0.3.0
Ruby Chemistry Development Kit (RCDK) version 0.3.0 is now available from RubyForge. RCDK enables the complete CDK API to be accessed from Ruby. This release adds support for IUPAC nomenclature translation and tighter Java integration.
Dependencies
RCDK requires Ruby, the Ruby developer libraries, a working build toolchain, and Ruby Java Bridge (RJB). This latter dependency can be satisfied during the RCDK installation process if the RubyGems method is used (see 'Installation').
Installation
RCDK can be conveniently installed using the RubyGems packaging mechanism:
sudo gem install rcdk
Alternatively, the source package and RubyGem can be downloaded here.
Tighter Java Integration
RCDK-0.3.0 introduces a previously-described Java package to Ruby module mapping mechanism. For example, if you'd like to create a Java ArrayList, it can be done through the new jrequire command:
require 'rubygems'
require_gem 'rcdk'
jrequire 'java.util.ArrayList'
list = Java::Util::ArrayList.new
list.size # => 0
IUPAC Nomenclature Translation
RCDK's most important new chemical informatics feature is made possible by Peter Corbett's excellent IUPAC nomenclature translation library OPSIN. It can either be used directly with jrequire, or indirectly through RCDK's convenience library RCDK::Util:
require 'rubygems'
require_gem 'rcdk'
require 'rcdk/util'
mol = RCDK::Util::Lang.read_iupac 'quinoline'
mol.getAtomCount # => 10
There are two things to notice here. First, no jrequire statement is needed when using the RCDK::Util library. Second, there is a multisecond delay after read_iupac is invoked. OPSIN itself introduces this delay during the NameToStructure constructor call, and RCDK inherits this behavior. However, after the first invocation of read_iupac, subsequent calls to this method are very fast.
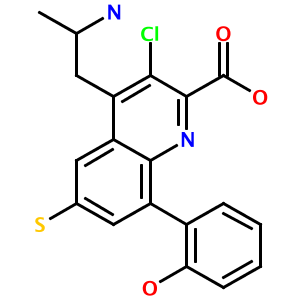
Let's decorate the quinoline nucleus with some substituents and render a 2-D image of the result. Execute the following code, either through the Ruby interpreter (ruby) or through Interactive Ruby (irb):
require 'rubygems'
require_gem 'rcdk'
require 'rcdk/util'
RCDK::Util::Image.iupac_to_png('3-chloro-4-(2-aminopropyl)-6-mercapto-8-(2-hydroxyphenyl)-quinoline-2-carboxylic acid', 'test.png', 300, 300)
Running this code produces the following image in your working directory:
Be Agile
RCDK marries the agility of the Ruby language with the functionality of three Open Source chemical informatics libraries: CDK; OPSIN; and Structure-CDK. Future articles will discuss some simple applications of this powerful combination.