From IUPAC Nomenclature to 2-D Structures With OPSIN
A previous article introduced OPSIN, an Open Source Java library for decoding IUPAC chemical nomenclature. In this tutorial, you'll see how OPSIN can, when interfaced with freely-available chemical informatics software, generate 2-D structure diagrams from IUPAC names.
Prerequisites
This tutorial requires Ruby CDK (RCDK), which in turn requires Ruby, Java, and the Ruby Java Bridge. Tutorials detailing the installation of RCDK on both Windows and Linux platforms are available.
In addition, you'll need a copy of the standalone jarfile opsin-big-0.1.0.jar. Future versions of RCDK will integrate the OPSIN jarfile, making this step unnecessary.
Outlining the Problem and a Solution
We'd like to create a simple Ruby class with a method that accepts an IUPAC chemical name as input and produces a PNG image of the corresponding molecule as output. OPSIN accepts IUPAC names as input, but it only produces Chemical Markup Language (CML) as output. The CML output lacks 2-D coordinates, and OPSIN itself has no 2-D rendering capabilities.
We'll use RCDK to augment OPSIN's capabilities. Thanks to CDK's built-in CML support, RCDK can read CML and generate an AtomContainer representation. RCDK also supports the assignment of 2-D coordinates to an AtomContainer via CDK's StructureDiagramGenerator. To produce the PNG image, we'll use the 2-D rendering capability made possible through Structure-CDK, which is a built-in component of RCDK.
A Simple Ruby Library
Create a working directory and copy opsin-big-0.1.0.jar into it. Next, create a file called depictor.rb containing the following Ruby code:
require 'rubygems'
require_gem 'rcdk'
require 'rcdk'
Java::Classpath.add('opsin-big-0.1.0.jar')
require 'util'
# A simple IUPAC->2-D structure convertor.
class Depictor
@@StringReader = import 'java.io.StringReader'
@@NameToStructure = import 'uk.ac.cam.ch.wwmm.opsin.NameToStructure'
@@CMLReader = import 'org.openscience.cdk.io.CMLReader'
@@ChemFile = import 'org.openscience.cdk.ChemFile'
def initialize
@nts = @@NameToStructure.new
@cml_reader = @@CMLReader.new
end
# Writes a width by height PNG to
# filename> for the molecule described by
# iupac_name.
def depict_png(iupac_name, filename, width, height)
cml = @nts.parseToCML(iupac_name)
throw("Can't parse name: #{iupac_name}") unless cml
molfile = cml_to_molfile(cml)
RCDK::Util::Image.molfile_to_png(molfile, filename, width, height)
end
private
def cml_to_molfile(cml)
string_reader = StringReader.new(cml.toXML)
@cml_reader.setReader(string_reader)
chem_file = @cml_reader.read(@@ChemFile.new)
molecule = chem_file.getChemSequence(0).getChemModel(0).getSetOfMolecules.getMolecule(0)
molecule = RCDK::Util::XY.coordinate_molecule(molecule)
RCDK::Util::Lang.get_molfile(molecule)
end
end
Testing, Testing
A short test will demonstrate the capabilities of the Depictor library. Add the following to a file called test.rb in your working directory (or enter it interactively with irb):
require 'depictor'
depictor = Depictor.new
name = '3,3-dimethyl-7-oxo-6-[(2-phenylacetyl)amino]-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid' #Penicillin G
depictor.depict_png(name, 'out.png', 300, 300)
Running this test produces a 300x300 PNG image of Penicillin G, named out.png, in your working directory:
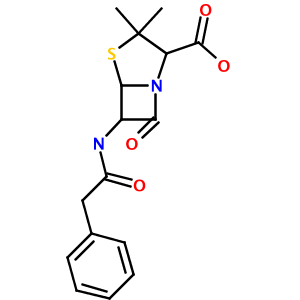
As you can see, this simple library and test code has:
- correctly parsed the rather complex IUPAC name (3,3-dimethyl-7-oxo-6-[(2-phenylacetyl)amino]-4-thia-1-azabicyclo[3.2.0]heptane-2- carboxylic acid) to a valid CML representation
- converted this representation to a CDK
AtomContainer - assigned 2-D coordinates
- rendered a PNG image in color
Notice how the thiaazabicyclo[3.2.0] system, complete with properly-placed substitutents, was flawlessly identified and parsed.
If you entered the above test code interactively via IRB, you may have noticed a multi-second delay in instantiating Depictor. This latency results from a sluggish NameToStructure constructor in OPSIN. A similar delay also occurs in OPSIN's pure-Java unit tests. Once Depictor is instantiated, however, image generation occurs relatively quickly.
The unususal orientation of the beta-lactam carbonyl group is determined by CDK's StructureDiagramGenerator. The source of this behavior will be explored in a future article.
More Examples
To illustrate some of the capabilities of the OPSIN-RCDK combination, a few more examples are provided below.
One of OPSIN's more surprising features is how well it handles heterocycles. For example, the IUPAC name for caffeine (1,3,7-trimethylpurine-2,6-dione) is translated to:
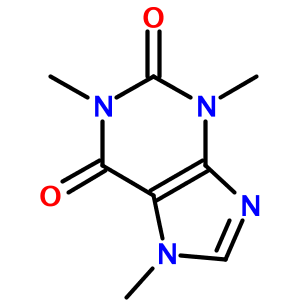
As another example, consider the tetrazole (1-[2-hydroxy-3-propyl-4-[3-(2H-tetrazol-5-yl)propoxy]phenyl]ethanone):
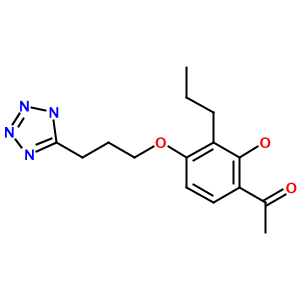
Highly substituted benzene rings and carboxylic acids are also translated accurately, as in 3-acetamido-5-(acetyl-methyl-amino)-2,4,6-triiodo-benzoic acid (Metrizoate):
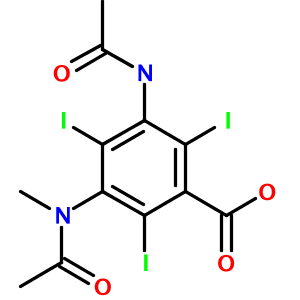
How about a hairy-looking macrocycle name with multiple levels of morpheme nesting (3,6-diamino-N-[[15-amino-11-(2-amino-3,4,5,6-tetrahydropyrimidin-4-yl)-8- [(carbamoylamino)methylidene]-2-(hydroxymethyl)-3,6,9,12,16-pentaoxo- 1,4,7,10,13-pentazacyclohexadec-5-yl]methyl]hexanamide)? Not a problem:
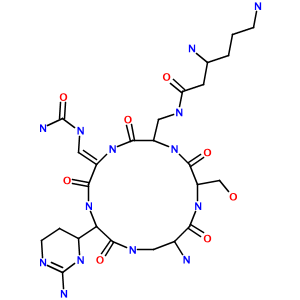
Limitations
In my tests of the OPSIN library, one structure appeared to be incorrectly parsed - N-(5-chloro-2-methyl-phenyl)-2-methoxy-N-(2-oxooxazolidin-3-yl)acetamide:
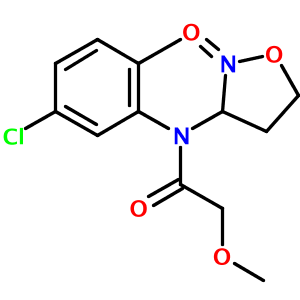
There are actually two problems with the output. First, an oxygen atom and a methyl group are overlapping near the top of the diargram. This cosmetic issue is related to CDK's StructureDiagramGenerator. Second, the oxazolidine nitrogen atom is misplaced by OPSIN. The correct 2-D image of this molecule, obtained from PubChem, is shown below:
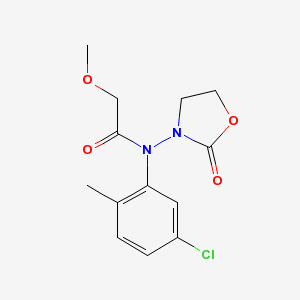
Conclusions
It's not common to find an early-development Open Source project with the sophistication of OPSIN. The smooth handling of nested morphemes, aromatic heterocycles, macrocycles, and a good fraction of what I threw at it leads me to belive that a well-designed and extensible nomenclature parsing engine lies at OPSIN's core. More on that later, though.
What could you do with a powerful Open Source IUPAC nomenclature parser? The answer to that one question could fill a three-volume series. Suffice it to say that OPSIN, in combination with other Open Source software, offers virtually limitless potential for indexing, collecting, repackaging, reprocessing, and mashing up vast amounts of chemical information. Because of its Open Source license, OPSIN can be extended and otherwise modified to fit your particular needs. Future articles will highlight some of the possibilities.