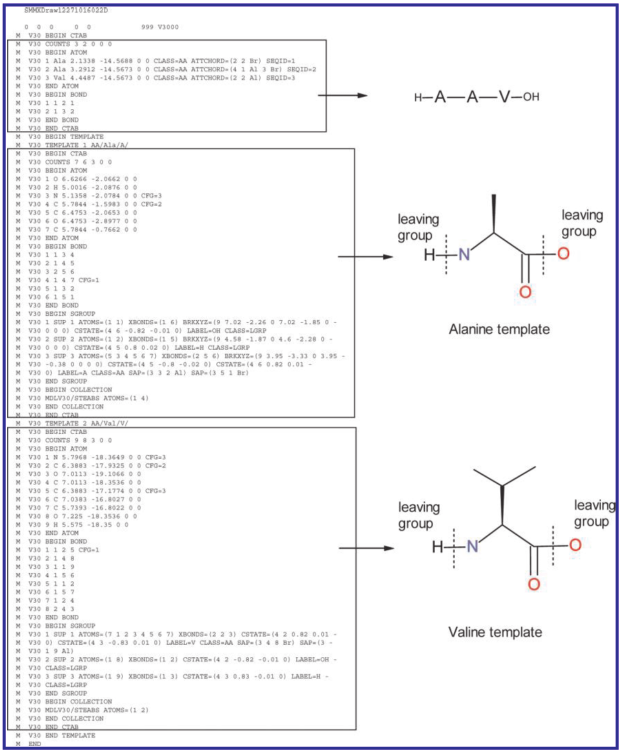
On the (F)utility of Extending the Molfile Format
MDL V2000 molfile format is the closest thing cheminformatics has to a universally-adopted standard. First publicly described in depth in 1991 and developed over the previous 13 years, the molfile format is read and written by nearly all software doing anything significant with organic chemical structures today.
Think about this for a moment - the lingua franca of cheminformatics predates Windows 3.0 and the World Wide Web. And it continues to be used as first described, with a few changes along the way.
Reliance on an old standard is not necessarily a bad thing. But when that standard's limitations hinder progress, it's time to look for alternatives. Has the time come to say 'goodbye' to the trusty V2000 molfile format, or does it still have plenty of life left?
V3000
A major limitation of V2000 is its hard cap on the number of atoms and bonds - 999. V2000 is a file format rooted in a bad idea long since abandoned, and made painfully obvious in the wake of Y2K - fixed-width fields. If your molecule contains more than 999 atoms or bonds you'll overflow the 'atom counts' field, and you'll be unable to specify a bond to the higher-numbered atoms.
Noting this and other problems, MDL (the originator of the V2000 spec) published a next-generation format called V3000 in 1995.
Over the course of the next fifteen years, V3000 went from merely being an option for large structures to the preferred format for encoding any structure, at least according to the molfile spec's authors. From the most recent June 2010 update:
Current Symyx products support reading and writing of both V2000 and V3000 formats. These products continue to default to writing V2000 molfiles to maximize interoperability with third party applications. Future product versions might default to output of the preferred V3000 format.
(Other changes were afoot as well: MDL was bought by Symyx, which was itself bought by Accelrys.)
Hard figures on the adoption of V3000 are difficult to come by. My sense in talking to others and reading the literature is that V3000 is being used internally by some large pharmaceutical companies - primarily for its ability to represent partially-defined stereochemistry.
However, when it comes to exchanging information between organizations, V2000 (in particular the V2000 Structure Data File) is the only game in town (SMILES files are a fallback, but that's a story for another time). For example, none of the Sixty-Four Free chemistry databases offering bulk downloads provide a V3000 option. They all use V2000.
Looking at the advantages stated by Symyx itself for V3000 - it's not hard to see why most in cheminformatics see little to be gained by switching:
- "Provides better support for new chemical properties or objects, and supports enhanced stereochemistry" [Needed by a minority in cheminformatics, and practical only under well-designed validation protocols.]
- "Removes fixed field widths to support large structures. …" [Cheminformatics has historically concerned itself with small molecules, so this has not been a major concern.]
- "Supports the use of templates in a template block, which is useful for representing large structures, such as biological molecules. …" [This is where things get interesting - see below].
- "Uses free format and tagging of information for easier parsing." [Unfortunately, V3000 was introduced before XML, so parsing these tags actually requires a lot of work by comparison.]
- "Provides better backward compatibility through BEGIN/END blocks." [V3000 attempts to offer a nested file structure, but again without the convenience of tooling available from a modern file format. This means more work for developers, not less.]
V3000 has so far failed to replace V2000 as an inter-organization exchange format. Furthermore, few software packages have fully implemented the V3000 spec (although the same could be argued for V2000 as well).
Is it possible that V3000, although solving some problems, didn't address enough of the right problems?
Self-Contained Sequence Representation
The pharmaceutical industry's innovation problems are no secret. In looking for solutions, one avenue gaining some traction is biopolymer-based drugs such as proteins, carbohydrates, and polynucleotides.
This has led to a new problem in cheminformatics - the need to effectively deal with co-mingled small molecule and biopolymer structures in corporate registration systems.
The problem is that cheminformatics has traditionally been concerned with small molecules - those typically having fewer than 100 heavy atoms. But it's not uncommon for biopolymers to consist of structures of thousands of heavy atoms or more. The optimizations that work so well for encoding, displaying, editing, and algorithmically processing large collections of small molecules begin to break down when applied to these larger molecules.
Very recently, Accelrys published their solution to this problem, Self-Contained Sequence Representation (SCSR). Based on the V3000 molfile format, SCSR offers a common framework for encoding both small molecules and biopolymers. In a whitepaper on this technology, Keith Taylor explains:
The biochemistry industry needs a method of representation for large structures that is meaningful to biologists and chemists, reduces redundant information and enables structural features to be searched using a computer system.
Remember those "templates" and "template blocks" touted as advantages for V3000 in the specification? This is where those data structures prove themselves.
In SCSR, atoms can define literal atomic species such as carbon or nitrogen. But atoms can also contain entire substructures, such as amino acid residues. Rather than repetitively encoding every atom in an alanine polypeptide residue, it's only necessary to give a pointer to a single instance of alanine encoded in the file - once per occurrence. This system saves considerable storage space, processing time, and leads to more more natural-looking depictions.
Chemically-modified residues and even polyvalent residues can both be handled using a consistent (if somewhat perplexing) notation system. Although focussed on biopolymers, this system could be adapted for use with any large molecule containing repeating subunits.
Points to note include:
- Self-Contained Sequence Representation is a V3000 extension only.
- Although fully implemented in Accelrys products, adoption of SCSR by major software vendors remains an open question at this point.
- SCSR is mainly a compression technology - it's possible to convert any V3000 SCSR file with complete fidelity into an expanded non-SCSR representation and vice versa.
It will be interesting to see to what extent the drive to unify chemical and biological registration systems moves more organizations away from V2000 molfiles. Supporting two file formats does require overhead that most groups would rather avoid. On the other hand, the need to support V2000 as an interchange format (e.g., chemical suppliers sending SD files to big pharmas) will likely hinder this transition.
V2000 Issues
Anyone who has worked with V2000 molfiles in even a moderate-sized structure database has been hit at last once with one of the technology's most serious shortcomings - V2000 molfiles can't faithfully encode a large amount of interesting small molecule chemistry.
Over the course of a few years, I detailed some of these cases on Depth-First. Many of the structures that can't be represented at sufficient detail involve multicenter bonding and axial chirality. Molecules like ferrocene and binapthalene are good representatives, but there are many others.
To be clear, these problems can be managed by applying ad-hoc fixes internally, but when it comes time for V2000 to play is star role as information exchange format, things can get ugly.
Zero-Order Bonds and Explicit Hydrogen Count Extension to V2000
Even simpler structures pose problems for V2000. Developing a solution by extending V2000 was the focus of a recent paper by Alex Clark.
V2000 provides two conveniences that have no doubt contributed to the widespread adoption of the format: (1) all bonds must be single, double, or triple; and (2) assignment of hydrogens to heavy atoms is optional. However, these conveniences also lead directly to some troubling cases of mistaken molecular identity.

Consider dimethyl tin and tin dichloride. The paper argues that file formats rooted in implicit hydrogen conventions make it impossible for software to determine how many hydrogens to assign to either species. It's implied that V2000 is such a format, and a proposed extension is detailed.
However, one of the lesser known features of the V2000 format is the atom valence field. In its typically terse way, the June 2010 release of CTfile Formats has this to say on the subject of the valence field:
Shows number of bonds to this atom, including bonds to implied H's.
Although sometimes considered a query-only feature, nothing in the V2000 specification supports this conclusion. On the contrary, V2000 defines another atom property called "hydrogen count" marked as "[Query]" (like bond topology and query bond types). The "valence" property is marked as "[Generic]" (like atom charge). Furthermore, V2000 supports an atom property in the properties block called "Substitution Count" that can be used with "[Query]" only and which does not override the atom "valence" property in the atom block (as other equivalent atom properties do). The terms "[Query]" and "[Generic]" are only ever defined in terms of the kinds of files that support them, so differences of opinion on this point are to be expected.
Regardless, the V2000 format apparently already provides a mechanism to distinguish tin(II) and tin(IV) oxidation states in the two examples above. The solution would therefore be for software encoders to specify a valence of "2" for tin(II) species and a valence of "4" for tin(IV) species, and for software decoders to respect this convention.
The extent to which software vendors support the atom valence attribute is an interesting question. A quick look at the CDK GitHub repository shows that MDLV2000Reader does in fact read the valence attribute, but this attribute is only assigned to instances of IPseudoAtom. The Open Babel GitHub repository shows no indication that the atom valence property is either read or written at all (mdlformat.cpp).
This is a little odd. The two most widely-used open source cheminformatics toolkits both ignore a built-in solution to the problem of implicit hydrogen counts of di- and tetravelent tin in V2000 molfiles. (Full disclosure - my company's structure editor ChemWriter also ignores the valence atom attribute). It seems likely that more than a few commercial packages take the same approach. Now a recent paper proposes an extension that would at least in part replicate the functionality of the atom valence attribute.
Another problem described in Clark's paper is the limitation of bond orders to one, two or three. There is no zero bond order. Take for example, the cobalt complex below:
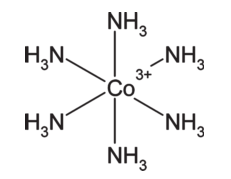
Cobalt is bonded to six amino groups, but in V2000 we're forced to give each bond a minimum order of 1. This leads to some strange conclusions. First, cobalt would have an implied formal charge of +9. Second, counting implicit hydrogens on each nitrogen atom (assuming we're not using the atom valence property) would lead to 2. We'd mis-calculate the molecular mass by 6 AMU. The paper considers some alternative representations, but each suffers from limitations of its own.
The paper proposes an extension to V2000 to allow for zero-order bonds, enabling the following improved representation to be displayed and used for calculations:
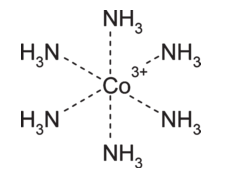
Although this solves the problem for certain classes of structure, it leaves others unsolved. For example, the zero-order bond representation of ferrocene below gives reasonably good calculations for formal charge, but it fails to capture the symmetry of the molecule, assigning significance to the two C-Fe bonds labeled as single where no significance is warranted and implying different bond types in each cyclopentyl ring, among other issues.
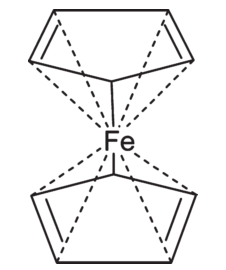
Finally, it's ironic that the proposed extensions apply to the much older V2000 format (the originator of which now claims will not be extended further). No mention is even made of the newer V3000 format that Accelrys says will be the basis of all future extensions.
Multiplication of Errors
V2000 is based on a very limited chemistry model, and this is the root cause of most of the problems cited by critics.
One of the most egregious errors is in the elevation of side-effects - atomic charge and bond order - to the status of fundamental atomic and bonding properties.
Ask any knowledgeable chemists how to calculate atomic formal charge and they'll do so by counting valence electrons. Valence electron count is the property any robust chemical interchange format should be capturing, not formal charge. Formal charge is the side-effect of a well-defined electron count. In case you missed that, let me re-state it:
Any chemical file format (or in-memory representation) that relies on charge as a fundamental atomic property is doomed to the same problems as V2000.
Bond order calculations, like formal charge calculations, are made by counting electrons. As a result, bond order is a very limiting property to choose as a basis for a chemical representation system. Bond order is derived from electron count, not the other way around. In other words:
Any chemical file format (or in-memory representation) that relies on bond order as a fundamental bond property is doomed to the same problems as V2000.
My concern is that attempts to extend the V2000 format by adding new templates (e.g., zero-order bond, and not to be confused with "templates" discussed above) will alleviate only part of the problem, but leave the rest festering. The problem with V2000 is not the lack of templates (e.g., no zero-order bond), but the use of templates like bond types in the first place.
Templates can also be found in stereochemical definitions, both in V2000 and elsewhere. All of the problems encountered with using V2000 to represent, for example axial chirality, can be traced to the reliance on templates.
Clark offers the following tongue-in-cheek witticism:
cheminformaticians do not know what molecule file format they will be using in 20 years, but they know it will be called MDL Molfile.
I would counter with:
cheminformaticians do not know what molecule file format will overcome the limitations of MDL Molfile, but they know it won't be based on templates.
Conclusions
No amount of retrofitting will solve the fundamental flaws of V2000 as a chemical representation and information exchange format. They are:
- Optional specification of implicit hydrogen counts.
- Encoding the side effect properties of charge and bond order, rather than the fundamental property of electron count.
- Elevation of bonding and stereochemistry templates to the status of fundamental properties.
- Deeply-ingrained and restrictive notions of bonding that precludes multi-center bonds or bonds with odd electron counts.
- Decades of inertia on the part of software providers and the cheminformatics community.
V3000 offers some true innovations in the areas of partial stereochemistry definition and biopolymer encoding that are worth understanding. However, at its core V3000 relies on the same basic (flawed) model of chemistry as V2000.
It's tempting to continue ignoring molecular corner cases as they accumulate. After all, if the only structures a cheminformatics system sees are small organic molecules, V2000 works nearly flawlessly. And if this is the only kind of molecule chemists continue to make, there really is no problem to solve.
But I'm optimistic about the future of chemical research and the creativity of chemists. It's time to retire both V2000 and V3000 molfile formats so that we in cheminformatics can stop making excuses and start keeping pace with advances in chemistry.