MX Performance Comparison #3: Substructure Search in MX and CDK
Substructure search is a fundamental cheminformatics operation. MX, the open source cheminformatics toolkit, contains an implementation based on the VF monomorphism algorithm. How fast is it? Let's compare it to CDK's UniversalIsomorphismTester.
The full report is available here. The full source code can be found on GitHub.
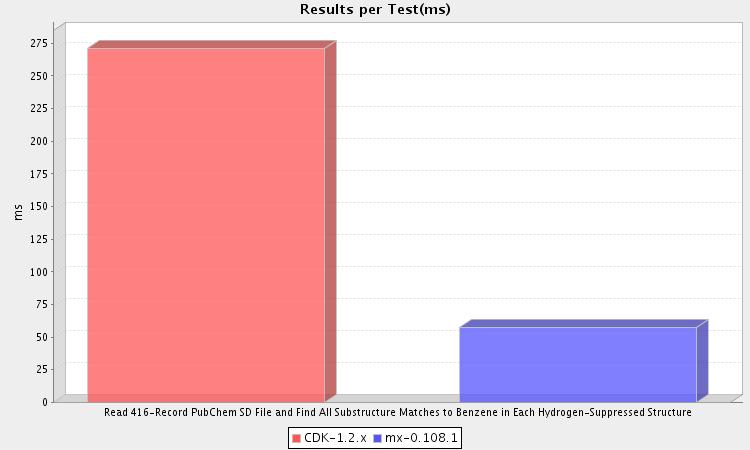
This test reads the molecules contained in a 416-record SD file into memory during setup. Then, during the test phase, each of these molecules is compared for a substructure relationship to a benzene molecule. As you can see, MX ran this test nearly five times faster than CDK.
MX and CDK differ in the algorithms used for substructure match. Whereas MX uses a variant of VF, CDK uses a variant of Ullmann. As noted by the VF creators, these two algorithms have very different performance characteristics, with VF always outperforming Ullmann. The performance gap increases quickly with increasing graph size.
CDK will soon have a new substructure matcher based on Rajarshi Guha's implementation. It will be interesting to directly compare this new CDK matcher to the one used by MX.
As is the case with the MX/CDK ring perception comparison, it should be noted that the MX substructure matcher implementation is optimized for readability and correctness, but not performance. A number of interesting opportunities exist for increasing the performance of the MX substructure matcher.
One point to note: MX and CDK differed in the number of hits found, with MX detecting all 416 and CDK finding 412. This is most likely due to the presence of isotopically-labelled benzenes in the dataset. Depending on your interpretation of a substructure match, either CDK or MX could be returning the "correct" answer.