Create Your Own PubChem Datasets - Exporting Results As SD Files
Recently, I needed to create a subset of the PubChem database in Structure Data File (SD File) format. Although it's far from obvious how to do this, the capability does exist. In this article, I'll give a step-by-step procedure for creating custom datasets in SD File format from arbitrary PubChem structure queries.
Create and Execute the Query
Let's say we want to create a dataset in SD File format containing all N-Boc-protected piperidines registered in PubChem.
From the main PubChem site, choose the Structure Search link. Then click the "Sketch" button.
Next, draw your molecule in the 2D structure editor:
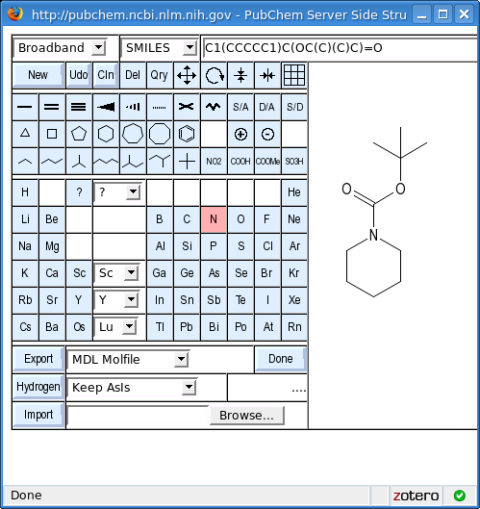
Then click the "Done" button.
Before starting the query (by clicking the "Search" button), be sure to select the "Substructure" option under "Search Type."
Exporting the Results
You should now be looking at a screen containing the first few hits of a 7700+ hitset. But how do we export these results in SD Format?
Next to a field labeled "Display", you'll see a drop-down box containing several different options. Choose the one labeled "PubChem Download."
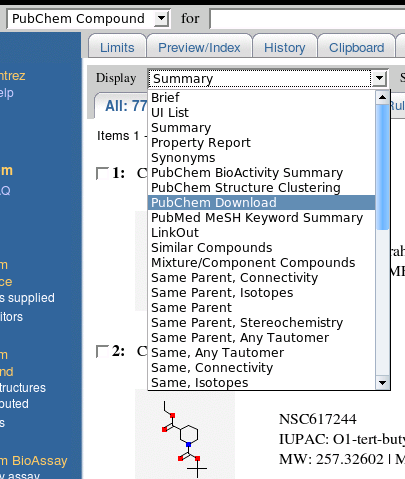
You'll be redirected to a download page from which you can select output formats, including SDF, or SD File. You can also select a compression type (datasets of even 2000 records can be quite large uncompressed). For this example, we'll select SDF format with GZip compression.
Clicking on the "Download" button takes us to a status page that eventually informs us when our download has been processed. You should then get a "Save File" dialog or something similar. If not, you should see a link to the compressed SD file.
Downloading the results file completes the process.