Never Draw the Same Molecule Twice: Image Metadata for Cheminformatics
The graphical language of 2D structures has served chemistry well for the last 100 years. Ironically, this language which is so useful for human communication is extraordinarily difficult for machines to understand. Heroic efforts at digital raster image recognition such as OSRA and those recently summarized by Egon Willighagen, in addition to a handful of others, have tried to tackle this problem with varying degrees of success.
The problem remains unsolved, and continues to be one of the most difficult technical challenges in cheminformatics. But the pace at which non-machine readable images are generated has accelerated dramatically in the last two years with the emergence of numerous free chemical databases.
What if 2D structure images simply contained all of the information needed for machine processing in the first place?
This idea isn't as far-fetched as it may sound initially. As discussed in a recent D-F article, both GChemPaint and ACD ChemSketch have been claimed to be capable of encoding machine-readable structure information.
Previous D-F articles have described "Firefly", the codename for a new lightweight 2D structure editor designed specifically for the Web. With major work on the editor's user interface complete, more recent efforts have focused on implementing a 2D rendering toolkit, and with it a mechanism to encode structural information within 2D molecular images.
As a demonstration of what is now possible, consider the structure of GlaxoSmithKline's diabetes treatment rosiglitazone (Avandia), depicted as a PNG image at the beginning of this article. At first glance, the image appears to be just like any other image of a 2D molecular structure. But it is not, for embedded within it are the connection table and 2D atom coordinates of rosiglitazone encoded as an industry-standard molfile.
Given the right software, a computer can interpret the structural information encoded in the rosiglitazone image and precisely re-create the original molecular representation. A graphical diagnostic tool bundled with Firefly was equipped with code for precisely this purpose.
This tool can work with molfile-encoded PNG images just as easily as it can with molfiles; they can be opened and the resulting molecule can be further edited, saved in another format, or re-written as a embedded-molfile PNG image.
The first step is to select the PNG image from a local hard drive:
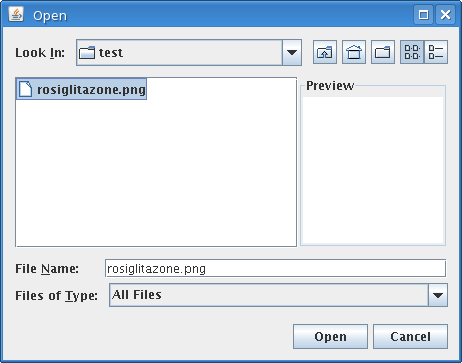
Opening this image produces a fully-editable version of the original molecule:
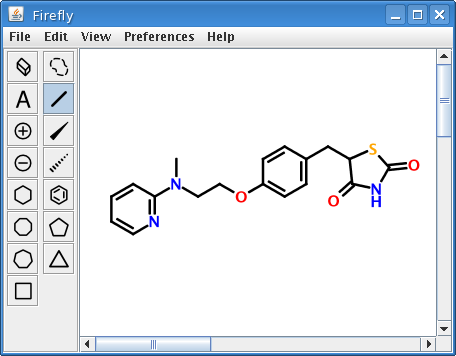
Obviously, nothing limits this technique to molfiles. InChI, SMILES, CML, or any other molecular encoding scheme would work just as well.
Using molecular-encoded PNG images as a Web-ready replacement for the Word/Chemdraw OLE technology may be one application of this approach. With a large corpus of these images, chemical Web spidering and data mining would be possible on a scale unimaginable today. As always, these possibilities reinforce the desperate need for high quality tools that chemists actually want to use, and which simultaneously yield machine-readable output.