Creating Canonical SMILES with Ruby Open Babel
Unlike many data types, molecular structure representations are not normally unique. Each numbering system you choose for the atoms and bonds of a molecule gives rise to completely accurate, but degenerate molecular representations. This is one of the fundamental peculiarities of chemical information - and the focus of much research activity over the last sixty or so years. One of the most widely-used approaches to this problem is canonicalization.
This article discusses the SMILES canonicalization capability in the upcoming Open Babel 2.1 release. Among several other enhancements, this release will also feature a brand new Ruby interface. By way of preview, this article will demonstrate just how convenient it has now become to generate canonical SMILES strings with Ruby.
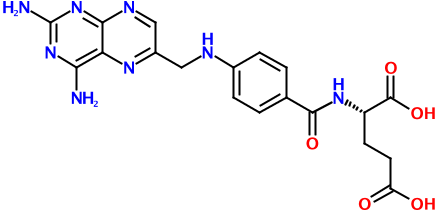
Consider the putative rodenticide aminopterin, the structure of which is shown above. Regardless of whether it turns out to be the culprit in the recent pet food poisoning case, it's a relatively complex molecule. And with this complexity comes many possible representations. Here's one of just hundreds, if not thousands, of possible SMILES strings for this molecule:
Nc3nc(N)c2nc(CNc1ccc(C(=O)N[C@@H](CCC(=O)O)C(=O)O)cc1)cnc2n3
If you were developing a database of molecules and needed to support exact structure searching, how would you do it? One way would be to convert a query molecule to a canonical SMILES string, and then simply look for that string in an index of your database's canonical SMILES. This is useful because it allows us to convert a chemistry-specific problem (exact structure search) into a generic computer science problem (text matching).
We can create a simple Ruby library to convert any SMILES string into an Open Babel canonical SMILES string:
require 'openbabel'
class Can
def initialize
@conversion = OpenBabel::OBConversion.new
@conversion.set_in_and_out_formats 'smi', 'can'
end
def convert smiles
mol = OpenBabel::OBMol.new
@conversion.read_string mol, smiles
@conversion.write_string mol
end
end
Save this code as a file called can.rb in your working directory. The library can then be used, for example, via interactive ruby (irb):
irb
irb(main):001:0> require 'can'
=> true
irb(main):002:0> c=Can.new
=> #<Can:0x2ac6cc653228 @conversion=#<OpenBabel::Conversion:0x2ac6cc6531d8>>
irb(main):003:0> puts c.convert('Nc3nc(N)c2nc(CNc1ccc(C(=O)N[C@@H](CCC(=O)O)C(=O)O)cc1)cnc2n3')
OC(=O)CC[C@@H](NC(=O)c1ccc(NCc2cnc3nc(N)nc(N)c3n2)cc1)C(=O)O
=> nil
irb(main):004:0> puts c.convert('C1=CC(=CC=C1C(=O)N[C@@H](CCC(=O)O)C(=O)O)NCC2=CN=C3C(=N2)C(=NC(=N3)N)N')
OC(=O)CC[C@@H](NC(=O)c1ccc(NCc2cnc3nc(N)nc(N)c3n2)cc1)C(=O)O
=> nil
Both SMILES strings for aminopterin were converted into the same canonical SMILES string.
Unlike InChI, which uses a "standard" canonicalization algorithm, SMILES canonicalization varies by software package. As a result, the SMILES canonicalization described here will be most useful within a software package, but probably not externally to it, at least initially.
Ruby is still an upstart language in cheminformatics. But tools like Ruby CDK and Ruby Open Babel offer ample opportunities for learning what this remarkable language can do for the development of chemistry applications.