Scripting Molecular Fingerprints with Ruby CDK
A molecular fingerprint represents a molecule as series of bits. There are many situations in which this reduced form of molecular representation is useful. For example, fingerprints are frequently used as a fast prescreen for database substructure searches. They can also be used for "fuzzy" comparisons involving molecular similarity, a nice complement to binary queries such as substructure search.
Fingerprints have their limitations. Being a form of hashing, they are imprecise in that two different molecules can have exactly the same fingerprint. The converse is also true: many molecular fingerprints exaggerate small differences between two molecules that most chemists would say are similar - for example between oxygen and sulfur analogs of the same structure.
Despite their limitations, the advantages of fingerprints make them useful in many situations. As a result, numerous fingerprinting systems have become popular. This tutorial will focus on creating and manipulating molecular fingerprints from Ruby using the Ruby Chemistry Development Kit (RCDK).
Prerequisites
For this tutorial, you'll need Ruby CDK (RCDK). A recent article described the small amount of system configuration required for RCDK on Linux. Another article showed how to install RCDK on Windows.
A Small Fingerprint Library
Let's build a small Ruby library for working with fingerprints. Place the following code into a file called fingerprint.rb in your working directory:
require 'rubygems'
require_gem 'rcdk'
require 'rcdk/util'
jrequire 'org.openscience.cdk.fingerprint.Fingerprinter'
jrequire 'org.openscience.cdk.similarity.Tanimoto'
# Molecule fingerprinting
class Fingerprinter
def initialize
@fingerprinter = Org::Openscience::Cdk::Fingerprint::Fingerprinter.new
end
def fingerprint(smiles)
mol = RCDK::Util::Lang.read_smiles smiles
fp = @fingerprinter.getFingerprint mol
# Metaprogramming!
fp.extend(Fingerprint)
end
end
# BitSet comparison
module Fingerprint
# Returns true of all of the bits set to true in this fingerprint are also set to true in the specified fingerprint
def subset?(fingerprint)
Org::Openscience::Cdk::Fingerprint::Fingerprinter.isSubset(fingerprint, self)
end
# Tanimoto similarity of this fingerprint and the specified fingerprint
def tanimoto(fingerprint)
Org::Openscience::Cdk::Similarity::Tanimoto.calculate(self, fingerprint)
end
end
Of particular note is the use of Ruby's Object.extend method. This method allows a single instance of an object to be extended at runtime - a form of metaprogramming. In this case, we add the subset? and tanimoto methods for determining whether all of the bits in one fingerprint are present in another, and for determining similarity, respectively. We use this technique here because currently RJB doesn't provide the complete interface into Java classes that would be required to create a Ruby class that directly inherits from Java's BitSet class.
Testing the Library
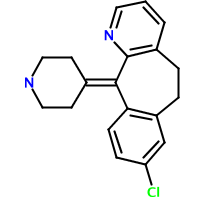
Claritin (loratadine, left) and Clarinex (desloratadine, right) are two structurally-related antihistamines. Can we quantitate the degree of similarity between these two structures? Fingerprints provide one way. The following code creates fingerprints for the two structures, determines if one is the subset of another, and assigns a Tanimoto similarity value:
require 'rubygems'
require_gem 'rcdk'
require 'rcdk/util'
jrequire 'org.openscience.cdk.fingerprint.Fingerprinter'
jrequire 'org.openscience.cdk.similarity.Tanimoto'
# Molecule fingerprinting
class Fingerprinter
def initialize
@fingerprinter = Org::Openscience::Cdk::Fingerprint::Fingerprinter.new
end
def fingerprint(smiles)
mol = RCDK::Util::Lang.read_smiles smiles
fp = @fingerprinter.getFingerprint mol
# Metaprogramming!
fp.extend(Fingerprint)
end
end
# BitSet comparison
module Fingerprint
# Returns true of all of the bits set to true in this fingerprint are also set to true in the specified fingerprint
def subset?(fingerprint)
Org::Openscience::Cdk::Fingerprint::Fingerprinter.isSubset(fingerprint, self)
end
# Tanimoto similarity of this fingerprint and the specified fingerprint
def tanimoto(fingerprint)
Org::Openscience::Cdk::Similarity::Tanimoto.calculate(self, fingerprint)
end
end
Variations
CDK's Fingerprinter class returns an instance of the Java class BitSet . This BitSet can be further manipulated in Ruby. For example, to find the size (the total number of bits) of the BitSet, we could use:
loratadine.size # => 1024
Similarly, to find the number of bits set to true, we would use:
loratadine.cardinality # => 278
To print out a list of all bits set to true, we could use the toString method:
loratadine.toString # => "{2, 8, 11, 16, 18, 22, 32, 37, 38, 41, 42, 46, 47, 51, 57, 64, 65, 66, 69 ... }"
Conclusions
Fingerprints enable many useful and fast comparisons between molecules. The form of fingerprint we've used here is but one of possibilities offered by CDK. The next article in this series will discuss fingerprints in Open Babel using both Ruby and Python.