Diversity-Oriented Chemical Informatics
How would you enumerate all of the molecules represented by a molecular formula? This question was recently posed to members of the Blue Obelisk mailing list. Formula-based exhaustive structure enumeration may seem on the surface to be just another esoteric problem. Nevertheless, playing with open, interactive software that can perform such enumerations can be a great source of new ideas for applications and unit tests.
The Chemistry Development Kit offers a fully-functional exhaustive structure enumerator through its GENMDeterministicGenerator class. This article will use GENMDeterministicGenerator through the Ruby CDK interface to generate color 2-D images for all molecules of a given molecular formula.
A Solution
The software described in this article will generate a collection of 2-D molecular PNG images based on a user-supplied molecular formula. When viewed in a file browser such as Windows Explorer or Konqueror, the output is visible as a matrix of images. The filename of each image is given by the SMILES string of the corresponding molecule. All molecules are enumerated, whether they look "reasonable" or not. As an example, consider a section of the output for 'C4H8ClNO', which looks like this on my system:
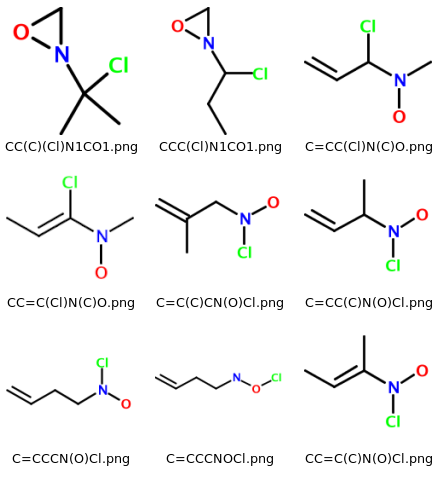
Enumerator: A Small Ruby Library
We'll create a small Ruby class to do most of the work. Save the following in a file called enum.rb:
require 'rubygems'
require_gem 'rcdk'
require 'rcdk/util'
jrequire 'org.openscience.cdk.structgen.deterministic.GENMDeterministicGenerator'
jrequire 'net.sf.structure.cdk.util.ImageKit'
class Enumerator
def initialize(formula)
@generator = Org::Openscience::Cdk::Structgen::Deterministic::GENMDeterministicGenerator.new(formula, '')
@width = 150
@height = 150
end
def set_size(width, height)
@width = width
@height = height
end
def write_images
mols = @generator.getStructures
iterator = mols.iterator
while (iterator.hasNext)
mol = RCDK::Util::XY.coordinate_molecule(iterator.next)
smiles = RCDK::Util::Lang.get_smiles(mol)
Net::Sf::Structure::Cdk::Util::ImageKit.writePNG(mol, @width, @height, "#{smiles}.png")
end
end
end
As you can see, this class is nothing more than a thin wrapper around a large amount of CDK functionality. Most of the action happens in the write_images method, where three things take place:
- We retrieve a list of molecules from the
GENMDeterministicGeneratorinstance that satisfy the molecular formula passed toEnumerator'sconstructor. - These molecules are iterated.
- For each molecule, an image is written with the filename given by its SMILES string.
Testing the Library
To test the library, the following code can either be entered interactively via Interactive Ruby (irb) or saved to a file and run with the Ruby interpreter (ruby):
require 'enum'
e=Enumerator.new 'C4H8ClNO'
e.write_images
Running this code will produce a collection of PNG images in your working directory. By changing the argument passed to the Enumerator constructor, you can change the makeup of the image set.
Prerequisites
For this tutorial, you'll need Ruby CDK (RCDK). A recent article described the small amount of system configuration required for RCDK on Linux. Another article showed how to install RCDK on Windows.
Unexpected Behavior
After testing the Enumerator library, you may notice a new file in your working directory called structuredata.txt. This file is written automatically by GENMDeterministicGenerator on instantiation, providing information on each structure that is generated. The CDK API does not mention the creation of this file, and it would be preferable for this file to only created on request. I'll be submitting a feature request to this effect shortly.
Food for Thought
If you plan to explore larger areas of chemical space with the Enumerator library, be prepared to wait. The generation of molecules, determination of 2-D coordinates, and rendering can take some time. Of course, the number of molecules increases dramatically with the number of atoms in the molecular formula - a concrete demonstration of what makes organic chemistry the fascinating discipline that it is.
An interesting variation on the ideas presented here would be to filter out molecules based on some criteria. One approach would be to remove molecules containing reactive functionality such as nitrogen substituted with chorine. A SMARTS pattern search could easily form the basis for this filter. In applying this and similar filters, larger areas of interesting chemical space could be sampled in a reasonable amount of time.
Conclusions
CDK's GENMDeterministicGenerator class, when combined with 2-D structure layout and 2-D rendering, provides the foundation of an intriguing tool for exploring chemical diversity. Further combining this capability with that offered by other freely-available tools offers some thought-provoking possibilities.